Abstract
It has been previously shown[1] that S. cerevisiae proteins preferentially interact with proteins of the same estimated likely time of origin. Using a similar approach but focusing on a less broad evolutionary time span I observed that the likelihood for protein interactions depends on the proteins’ age.
Methods and Results
Protein-protein interactions for S. cerevisiae were obtained from BIND, excluding any interactions derived from protein complexes. I considered only proteins that were represented in this interactome (i.e. with one or more interactions).
In order to create groups of S. cerevisiae proteins with different average age I used the reciprocal best blast hit method to determine the most likely ortholog in eleven other yeast species (see figure 1 for species names).
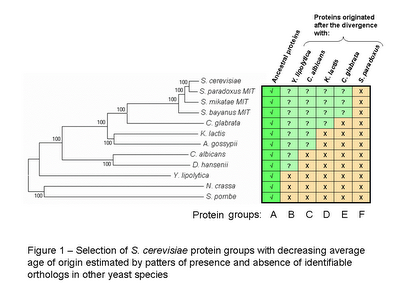
S. cerevisiae proteins with orthologs in all species were considered to be ancestral proteins and were grouped into group A. To obtain groups of proteins with decreasing average age of origin, S. cerevisiae proteins were selected according to the absence of identifiable orthologs in other species (see figure 1). It is important to note that these groups of decreasing average protein age are overlapping. Group F is contained in E , both are contained in D and so forth. I could have selected non overlapping groups of proteins with decreasing time of origin but the lower numbers obtained might in a latter stage make statistical analysis more difficult.
The phylogenetic tree in figure 1 (obtained with MEGA 3.1) is a neighbourhood joining tree obtained by concatenating 10 proteins from the ancestral group A. I did it mostly to avoid copyrighted images and too have a graphical representation of the species divergence.
To determine the effect of protein age on the likelihood of interaction with ancestral proteins I counted the number of interactions between group A and the other groups of proteins (see table 1).
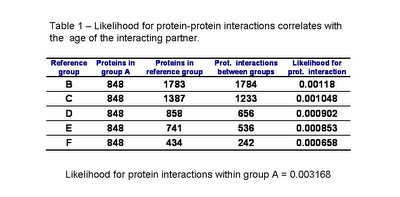
From the data it is possible to observe that protein-interactions within groups (within group A) is more likely than protein-interactions between groups. This is in agreement with the results from Qin et al.[1]. Also the likelihood for a protein to interact with an ancestral protein depends on the age of this protein. This simple analysis suggests that the younger the protein is the less likely it is to interact with an ancestral protein.
One possible use of this observation, if it holds to further scrutiny, would be to use the likely time of origin of the proteins as information to include in protein-protein prediction algorithms.
Caveats and possible continuations
The protein-protein interactions used here also contain the high-throughput studies and therefore the interactome used should be considered with caution. I might redo this analysis with a recent set of interactions compiled from the literature[2] but this will also introduce some bias into the interactome.
I should do some statistical analysis to determine if the differences observed are at all significant. If the differences are significant I should try to correlate the likelihood of interactions with a quantitative measure like average protein identity.
References
[1]Qin H, Lu HH, Wu WB, Li WH. Evolution of the yeast protein interaction network. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12820-4. Epub 2003 Oct 13
[2]Reguly T, Breitkreutz A, Boucher L, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006 Jun 8;5(4):11 [Epub ahead of print]
Tags: