System Biology quick links
(via Pierre) BMC System Biology has published their first papers. More or less at the same time the new Systems and Synthetic Biology (published by Springer Netherlands) has started publishing papers. These two journals join IEE Systems Biology and Molecular Systems Biology (Nature/EMBO) as forums to publish works on Systems and Synthetic Biology. All journals (with the exception of IEE Systems Biology) publish in open access or at least (in the case of Systems and Synthetic Biology) offer an open access option.
Some of the talks from the BioSysBio conference are online in Goggle Video.
Here is a nice talk from Alfonso Valencia talking about species co-evolution and a very promising improvement to a sequence based method to predict protein-protein interactions:
Tuesday, January 23, 2007
Monday, January 22, 2007
Social gene annotation in Connotea
There has been a lot of excitement over the recent web technological developments. Time magazine has recognized this by announcing that instead of profiling an individual in their annual issue of Person of Year they decided to select You as the most influential group of last year. This "you" refers to everyone that is out there on the web building, interacting, blogging, uploading their videos and pictures for the world to see. As with almost every rising meme, the backlash is inevitable. Some see this web euphoria as little more than global narcissism.
This social web holds some powerful promises of more efficient collaboration but clear examples might still be lacking. Scientists, given our need to communicate and collaborate, are a group of individuals that could do more to take advantage of these tools. Unfortunately we seem to be too unaware and too slow to pick them up.
I have shown before that the accumulating body of knowledge in Connotea, in the form of simple tagging of science papers, can in principle be used to highlight papers of higher impact.
I tough that it could also be possible to mine Connotea to retrieve gene annotations. I tested if manuscripts tagged as "cell-cycle" and "yeast" would contain, in their abstracts, mostly genes names related to cell cycle in yeast. There are currently 38 papers in Connotea tagged as cell-cycle and yeast with an associated Pubmed ID. I used a dictionary of S. cerevisiae gene names obtained from SGD and retrieved the abstracts for the 38 manuscripts using eUtils.
Within these abstracts there were 38 gene names associated by a simple pattern match. To evaluate the performance of this social gene annotation I took from the SGD's slim GO mapping the function and processes associated to these genes. I also included the gene description from gene name registry.
Table 1 - Known GO process/function annotations and gene function description associated to the genes predicted to participate in cell-cycle in yeast by social annotations.
From the 38 genes, 14 (~37%) are annotated in the slim GO annotation as participating in cell-cycle,meiosis or cytokinesis. From the remaining, 15 (39%) have a described function associated to the cell-cycle (ex. G1 cyclin involved in cell cycle progression, expression restricted to mother cells in late G1 as controlled by Swi4p-Swi6p, Swi5p and Ash1p,etc). In total roughly 76% of the gene names obtained are associated to cell-cycle in S. cerevisiae.

This simple test highlights the potential usefulness of social bookmarking of science papers. However it was limited to a very specific field and to a very small number of annotated manuscripts. Hopefully someone can come up with a better way of testing this :).
There has been a lot of excitement over the recent web technological developments. Time magazine has recognized this by announcing that instead of profiling an individual in their annual issue of Person of Year they decided to select You as the most influential group of last year. This "you" refers to everyone that is out there on the web building, interacting, blogging, uploading their videos and pictures for the world to see. As with almost every rising meme, the backlash is inevitable. Some see this web euphoria as little more than global narcissism.
This social web holds some powerful promises of more efficient collaboration but clear examples might still be lacking. Scientists, given our need to communicate and collaborate, are a group of individuals that could do more to take advantage of these tools. Unfortunately we seem to be too unaware and too slow to pick them up.
I have shown before that the accumulating body of knowledge in Connotea, in the form of simple tagging of science papers, can in principle be used to highlight papers of higher impact.
I tough that it could also be possible to mine Connotea to retrieve gene annotations. I tested if manuscripts tagged as "cell-cycle" and "yeast" would contain, in their abstracts, mostly genes names related to cell cycle in yeast. There are currently 38 papers in Connotea tagged as cell-cycle and yeast with an associated Pubmed ID. I used a dictionary of S. cerevisiae gene names obtained from SGD and retrieved the abstracts for the 38 manuscripts using eUtils.
Within these abstracts there were 38 gene names associated by a simple pattern match. To evaluate the performance of this social gene annotation I took from the SGD's slim GO mapping the function and processes associated to these genes. I also included the gene description from gene name registry.
Table 1 - Known GO process/function annotations and gene function description associated to the genes predicted to participate in cell-cycle in yeast by social annotations.
From the 38 genes, 14 (~37%) are annotated in the slim GO annotation as participating in cell-cycle,meiosis or cytokinesis. From the remaining, 15 (39%) have a described function associated to the cell-cycle (ex. G1 cyclin involved in cell cycle progression, expression restricted to mother cells in late G1 as controlled by Swi4p-Swi6p, Swi5p and Ash1p,etc). In total roughly 76% of the gene names obtained are associated to cell-cycle in S. cerevisiae.

This simple test highlights the potential usefulness of social bookmarking of science papers. However it was limited to a very specific field and to a very small number of annotated manuscripts. Hopefully someone can come up with a better way of testing this :).
Thursday, January 18, 2007
Petition for guaranteed public access
(via PLoS publishing blog):
"A group of European organisations - JISC (Joint Information Systems Committee, UK), SURF (Netherlands), SPARC Europe, DFG (Deutsches Forschungsgemeinschaft, Germany), DEFF (Denmark's Electronic Research Library) - have posted a petition to encourage the EC to formally endorse the open access recommendations."
This petition recommends that "any potential 'embargo' on free access should be set at no more than six months following publication" for any EC funded research.
Have a look and sign the petition if you are for it.
(via PLoS publishing blog):
"A group of European organisations - JISC (Joint Information Systems Committee, UK), SURF (Netherlands), SPARC Europe, DFG (Deutsches Forschungsgemeinschaft, Germany), DEFF (Denmark's Electronic Research Library) - have posted a petition to encourage the EC to formally endorse the open access recommendations."
This petition recommends that "any potential 'embargo' on free access should be set at no more than six months following publication" for any EC funded research.
Have a look and sign the petition if you are for it.
Sunday, January 14, 2007
Bio::Blogs# 7 and some quick links
The bioinformatics blog journal Bio::Blogs will have it's 7th edition on the 1st of February. We skipped the January edition because of the holidays. It will be hosted by Paras Chopra on BioHacking blog. Anyone can submit the link to their posts on paras1987 {at} gmail or bioblogs {at} gmail until the end of this month.
Some quick links:
Paras released the source code of a Python program for protein structure prediction.
(via Gerstein' blog) Yale university has a podcast. I wish I could convince EMBL's press office to start blogging and/or a podcast.
(via Deepak) The Science Commons blog announced that the three journals published by EMBO and NPG (EMBO reports, EMBO journal and Molecular Systems Biolgoy) will soon start publishing with a creative commons license. More information on the subject can be found in the EMBO site. In the case of Molecular Systems Biology all articles are published in open access but for EMBO Journal and EMBO reports it looks like the author will decide if they wish to pay an extra fee (2000 euros) to publish in open access. Only the articles published in open access will be published with the creative commons license. Adopting the creative commons license will make re-using their papers much easier, hopefully increasing the usefulness of their content.
(disclaimer: I am currently working for Molecular Systems Biology. All opinions expressed in this blog are my own)
Speaking of re-using content. Alf has set up a mirror site for PLoS One. He called it PLoS Too :) and he is using it to try out some ideas on layout, microformats and features like rating. This is one funny thing about the creative commons license. As long as you give credit to the source you are free to re-use the content. Nothing stops a group of people from setting up a new journal, based on those that are published in creative commons, with a different editorial line. For this particular license you can even try to make some money from re-using the content :).
The bioinformatics blog journal Bio::Blogs will have it's 7th edition on the 1st of February. We skipped the January edition because of the holidays. It will be hosted by Paras Chopra on BioHacking blog. Anyone can submit the link to their posts on paras1987 {at} gmail or bioblogs {at} gmail until the end of this month.
Some quick links:
Paras released the source code of a Python program for protein structure prediction.
(via Gerstein' blog) Yale university has a podcast. I wish I could convince EMBL's press office to start blogging and/or a podcast.
(via Deepak) The Science Commons blog announced that the three journals published by EMBO and NPG (EMBO reports, EMBO journal and Molecular Systems Biolgoy) will soon start publishing with a creative commons license. More information on the subject can be found in the EMBO site. In the case of Molecular Systems Biology all articles are published in open access but for EMBO Journal and EMBO reports it looks like the author will decide if they wish to pay an extra fee (2000 euros) to publish in open access. Only the articles published in open access will be published with the creative commons license. Adopting the creative commons license will make re-using their papers much easier, hopefully increasing the usefulness of their content.
(disclaimer: I am currently working for Molecular Systems Biology. All opinions expressed in this blog are my own)
Speaking of re-using content. Alf has set up a mirror site for PLoS One. He called it PLoS Too :) and he is using it to try out some ideas on layout, microformats and features like rating. This is one funny thing about the creative commons license. As long as you give credit to the source you are free to re-use the content. Nothing stops a group of people from setting up a new journal, based on those that are published in creative commons, with a different editorial line. For this particular license you can even try to make some money from re-using the content :).
Thursday, January 11, 2007
Scientific Journals blog
Blogs have been around for some time. From the wikepedia:
Blogs in science, on the other hand, have only recently become popular. The first two traditional science news journals to pick up the raising interest in scientific blogging were The-Scientist in August 2005, and then Nature in December 2005. Since then many more scientist have picked up blogging for a variety of purposes (see review by Coturnix). The science journals have been slowly reacting. Here is a current list of blogs from science journal blogs or publishing groups. If anyone knows more please leave a comment and I will add them to the list.
Blogs have been around for some time. From the wikepedia:
The term "weblog" was coined by Jorn Barger on 17 December 1997. The short form, "blog," was coined by Peter Merholz, who jokingly broke the word weblog into the phrase we blog in the sidebar of his blog Peterme.com in April or May of 1999.
Blogs in science, on the other hand, have only recently become popular. The first two traditional science news journals to pick up the raising interest in scientific blogging were The-Scientist in August 2005, and then Nature in December 2005. Since then many more scientist have picked up blogging for a variety of purposes (see review by Coturnix). The science journals have been slowly reacting. Here is a current list of blogs from science journal blogs or publishing groups. If anyone knows more please leave a comment and I will add them to the list.
| Journal | Blog |
| The Scientist | |
| Scientific American | |
| Nature publishing group | |
| Nature Journals | |
| Nature Genetics | |
| Nature Neuroscience | |
| Nature Methods | |
| Nature Medicine | |
| Nature News | |
| Chemistry at Nature (portal not journal) | |
| Nature publishing group | |
| Nature publishing group | |
| Nature publishing group | |
| Heredity | |
| Public Library of Science | |
| PLoS Blogs | |
| PLoS publishing | |
| PLoS Technology | |
| PLoS Medicine | |
| The Lancet | |
| Science | The Weblog of Science Magazine"s (stopped) |
Wednesday, January 10, 2007
Science Blogging Anthology 2006
I mentioned before that Coturnix was getting ready a list of blog posts that would go into a science blogging anthology of 2006. Twelve judges have selected 50 blog posts that will be put together in a book. The book will then be published by lulu. The judges were nice enough to select one of my posts to go in the book :)
Opening up the scientific process
I mentioned before that Coturnix was getting ready a list of blog posts that would go into a science blogging anthology of 2006. Twelve judges have selected 50 blog posts that will be put together in a book. The book will then be published by lulu. The judges were nice enough to select one of my posts to go in the book :)
Opening up the scientific process
Saturday, January 06, 2007
Science Blogging Anthology
Coturnix from a Blog Around the Clock is organizing a science blogging anthology. I missed it during the Christmas holidays but the results are due in a couple of days. Here is the list of posts that got nominated and are now being evaluated. One of my posts made the nomination list :) cool.
Last month Roland Krause said that this type of vanity posts (like blog carnivals) are similar to spam blogs. I actually think that there is value in carnivals and other equivalent content promotion activities. They create a cheap reward system that motivates people to produce more and better content. They also provide with a layer of quality rating even if, in the case of carnivals, the posts that are submitted are self contributed. Bloggers tend to submit their best content to the carnivals.
On a related note but with a very different opinion, here is a rant on Web 2.0 And Narcissism (via Rough Type):
Coturnix from a Blog Around the Clock is organizing a science blogging anthology. I missed it during the Christmas holidays but the results are due in a couple of days. Here is the list of posts that got nominated and are now being evaluated. One of my posts made the nomination list :) cool.
Last month Roland Krause said that this type of vanity posts (like blog carnivals) are similar to spam blogs. I actually think that there is value in carnivals and other equivalent content promotion activities. They create a cheap reward system that motivates people to produce more and better content. They also provide with a layer of quality rating even if, in the case of carnivals, the posts that are submitted are self contributed. Bloggers tend to submit their best content to the carnivals.
On a related note but with a very different opinion, here is a rant on Web 2.0 And Narcissism (via Rough Type):
"What he's getting at is that this whole Web 2.0, social networking, virtual community business is essentially a pornography of the self—a projected, fictionalized self that is then worshipped by the slightly less-perfect self."
Thursday, January 04, 2007
Specificity and Evolvability in Protein Interaction Networks
I finally have the opportunity to blog about some of what I have worked on during last year. It has been published in PLoS Computational Biology and is freely available here in the early online release format (still in the original ugly format :). One way to use our blogs may be to add some depth to the papers that we published. Something like the extras we get when we buy the DVD of a movie ;)
Main conclusions
- Protein interactions can change at a fast rate of 1E-5 interactions per protein pair per million years
- Binding specificity is a strong determining of binding specificity with more promiscuous binding proteins having a higher rate of change of interactions.
- Human proteins involved in immune response, transport and establishment of localization, show signs of positive selection for change of interactions.
The making of
We had been using comparative genomics to search for conserved putative protein binding sites. These very conserved putative target sites were very likely to be experimentally known target sites but many other known binding sites seamed not to be so conserved. This was what got us started thinking about the evolution of these protein interactions and what might determine the rate at which interactions are gained and lost during evolution. The analysis was mostly inspired on the nice work of Andreas Wagner that first proposed a rate for the addition of new interactions in S. cerevisiae. We have tried to build on this by analyzing different species and determining also what protein properties might determine the rate at which interactions are gained and lost in evolution.
More than nodes and edges
One of the main conclusions from this work was that binding specificity also determines the rate of change of interactions during evolution. More promiscuous binding proteins not only have many binding partners but they also tend to change partners faster during evolution. To establish a proxy for binding specificity we have used structural information from the iPFAM database. In essence we have considered that protein domains that have been seen in contact with many different other domains would be more promiscuous. In general we observed that proteins containing these promiscuous domains had a high rate of change of interactions (see figure below).
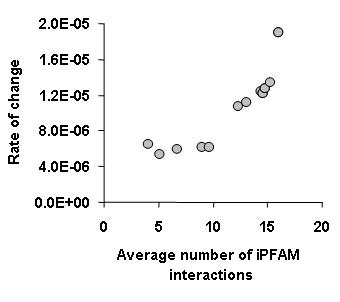
This highlights something that I had stressed before, that it is important to consider protein interaction networks as more than nodes and edges. Another recent paper has also shown that it is possible and useful to use the accumulating structural information available in the PDB to obtain a more accurate representation of protein interaction networks. Philip M. Kim and co workers from the Gerstein lab (blog, webpage) published a study in Science were they have used also the iPFAM database to curate all the S. cerevisie interactions and to discriminate between interactions that use the same or different binding interfaces. With this information the authors distinguish between hubs that tend to interact with their partners mostly trough one interface or trough many interfaces. They have shown that the multi interface hubs are more restricted in evolution and more likely to be essential than single interface hubs. They have a website with presentations and additional data for this paper.
In the pipeline
To be submitted soon (hopefully), are some collaborations on how to use structural information to predict protein-protein binding specificity. With these collaborations I finish my thesis (still waiting for the defense). During the next couple of months I am off to search for a lab to work as a postdoc.
I finally have the opportunity to blog about some of what I have worked on during last year. It has been published in PLoS Computational Biology and is freely available here in the early online release format (still in the original ugly format :). One way to use our blogs may be to add some depth to the papers that we published. Something like the extras we get when we buy the DVD of a movie ;)
Main conclusions
- Protein interactions can change at a fast rate of 1E-5 interactions per protein pair per million years
- Binding specificity is a strong determining of binding specificity with more promiscuous binding proteins having a higher rate of change of interactions.
- Human proteins involved in immune response, transport and establishment of localization, show signs of positive selection for change of interactions.
The making of
We had been using comparative genomics to search for conserved putative protein binding sites. These very conserved putative target sites were very likely to be experimentally known target sites but many other known binding sites seamed not to be so conserved. This was what got us started thinking about the evolution of these protein interactions and what might determine the rate at which interactions are gained and lost during evolution. The analysis was mostly inspired on the nice work of Andreas Wagner that first proposed a rate for the addition of new interactions in S. cerevisiae. We have tried to build on this by analyzing different species and determining also what protein properties might determine the rate at which interactions are gained and lost in evolution.
More than nodes and edges
One of the main conclusions from this work was that binding specificity also determines the rate of change of interactions during evolution. More promiscuous binding proteins not only have many binding partners but they also tend to change partners faster during evolution. To establish a proxy for binding specificity we have used structural information from the iPFAM database. In essence we have considered that protein domains that have been seen in contact with many different other domains would be more promiscuous. In general we observed that proteins containing these promiscuous domains had a high rate of change of interactions (see figure below).

This highlights something that I had stressed before, that it is important to consider protein interaction networks as more than nodes and edges. Another recent paper has also shown that it is possible and useful to use the accumulating structural information available in the PDB to obtain a more accurate representation of protein interaction networks. Philip M. Kim and co workers from the Gerstein lab (blog, webpage) published a study in Science were they have used also the iPFAM database to curate all the S. cerevisie interactions and to discriminate between interactions that use the same or different binding interfaces. With this information the authors distinguish between hubs that tend to interact with their partners mostly trough one interface or trough many interfaces. They have shown that the multi interface hubs are more restricted in evolution and more likely to be essential than single interface hubs. They have a website with presentations and additional data for this paper.
In the pipeline
To be submitted soon (hopefully), are some collaborations on how to use structural information to predict protein-protein binding specificity. With these collaborations I finish my thesis (still waiting for the defense). During the next couple of months I am off to search for a lab to work as a postdoc.
Monday, December 25, 2006
Publish your GreaseMonkey scripts
I knew it was possible to use GreaseMonkey scripts to change webpages to better suit our needs. I tried it once to get blog comments about scientific papers to show up in journal websites. Pierre and Stew have been creating several interesting scripts for postgenomic and connotea. What I did not know was that one could actually publish these scripts. I am all for publishing of smaller and finner grained scientific content but I have to say that this paper seemed like little more than a big blog post. Should we try to publish this type of work ?
Anyway :) ho ho ho , merry xmas everyone
I knew it was possible to use GreaseMonkey scripts to change webpages to better suit our needs. I tried it once to get blog comments about scientific papers to show up in journal websites. Pierre and Stew have been creating several interesting scripts for postgenomic and connotea. What I did not know was that one could actually publish these scripts. I am all for publishing of smaller and finner grained scientific content but I have to say that this paper seemed like little more than a big blog post. Should we try to publish this type of work ?
Anyway :) ho ho ho , merry xmas everyone
Friday, December 22, 2006
PLoS One is up and running
I will add this blog's "voice" to the many that have already announced the start of PLoS One. This journal is a very much needed experiment in science communication online. It is being built from scratch to take advantage of the internet as the medium unlike many other journals. As Deepak mentioned in his blog, the success of One will depend mostly on us, the users, in our interest to participate with our own insight. The PLoS One team will have to worry about creating interesting reward systems around the journal to help boost participation.
I have only played around on the site for a short while but there are a couple of features that I hope that will implement in the short term.
- One that I was hoping to find at start was some form of gateways or portals for areas. The only subject navigation available seams to be the links on the right side.
- I also did not find any kind of rating system. To boost participation I think people have to start trying out simple participation systems and rating is the easiest one.
- I guess it would also be nice to have some kind of track back system or some other way to let comments come in from blog post. This would be nice for bloggers but i have to admit that few people would care :) .
There were two papers (from the same group) that caught my attention but I did not have time to read them:
Control of Canalization and Evolvability by Hsp90
Modularity and Intrinsic Evolvability of Hsp90-Buffered Change
I will add this blog's "voice" to the many that have already announced the start of PLoS One. This journal is a very much needed experiment in science communication online. It is being built from scratch to take advantage of the internet as the medium unlike many other journals. As Deepak mentioned in his blog, the success of One will depend mostly on us, the users, in our interest to participate with our own insight. The PLoS One team will have to worry about creating interesting reward systems around the journal to help boost participation.
I have only played around on the site for a short while but there are a couple of features that I hope that will implement in the short term.
- One that I was hoping to find at start was some form of gateways or portals for areas. The only subject navigation available seams to be the links on the right side.
- I also did not find any kind of rating system. To boost participation I think people have to start trying out simple participation systems and rating is the easiest one.
- I guess it would also be nice to have some kind of track back system or some other way to let comments come in from blog post. This would be nice for bloggers but i have to admit that few people would care :) .
There were two papers (from the same group) that caught my attention but I did not have time to read them:
Control of Canalization and Evolvability by Hsp90
Modularity and Intrinsic Evolvability of Hsp90-Buffered Change
Tags: plos,plosone,open science
Tuesday, December 05, 2006
Second (hellish) Life
I had a walk around Second Nature inside Second Life. I had tried SL before some months ago and this time the experience was much worse. My avatar keeps freezing when I touch objects or when I stand around idle for some time. Some other times the avatar just freezes in flight and continues in the same direction until I quit. It was just unusable.
Here is a picture from an 3D cell in the Second Nature island.

A feeble attempt to build something for the EMBL online symposium

I had a walk around Second Nature inside Second Life. I had tried SL before some months ago and this time the experience was much worse. My avatar keeps freezing when I touch objects or when I stand around idle for some time. Some other times the avatar just freezes in flight and continues in the same direction until I quit. It was just unusable.
Here is a picture from an 3D cell in the Second Nature island.

A feeble attempt to build something for the EMBL online symposium

Tags: second life,science
Monday, December 04, 2006
Modularity and Evolvability
The First Online EMBL PhD Symposium as started today with several media files available for viewing and commenting. There is an IRC channel open for discussion.
Participants can comment or add their own content to the site. I have put up a small presentation relating modularity and evolvability in proteins, language, software and the scientific process. I am no expert in any of these fields so view these analogies with a very critical eye :).
Full screen view can be access from the slideshare site
The First Online EMBL PhD Symposium as started today with several media files available for viewing and commenting. There is an IRC channel open for discussion.
Participants can comment or add their own content to the site. I have put up a small presentation relating modularity and evolvability in proteins, language, software and the scientific process. I am no expert in any of these fields so view these analogies with a very critical eye :).
Full screen view can be access from the slideshare site
Saturday, December 02, 2006

The 6th edition of Bio::Blogs is up in Nodalpoint. Many thanks to Greg for setting it up. This edition marks half a year of Bio::Blogs and it is dedicated to conference blogging, probably one of the best examples of the usefulness of science blogging.
The 7th edition will probably be scheduled to February to skip the holiday season and Paras Chopra has volunteered to host it.
Tags: bio::blogs,blog canivals,bioblogs
Monday, November 27, 2006
EMBL online PhD symposium

(via Notes from the biomass)
EMBL is organizing the 1st PhD online symposium (4-8 December). It will be fully online, available for free to anyone. Participants can register to participate in the discussions and watch the presentations when they are made available. The users can create pages to share posters, talks or anything that might be relevant to the conference. In all, it could be a good playground to try out new ideas for online conferences. We could ask Nature to broadcast it in Nature island inside second life ? :)
Register, create your pages, share some thoughts. The topics are:
Career Development
Omics Session / Systems Biology
Scientific Communication 2.0
Konrad is one of the organizers.

(via Notes from the biomass)
EMBL is organizing the 1st PhD online symposium (4-8 December). It will be fully online, available for free to anyone. Participants can register to participate in the discussions and watch the presentations when they are made available. The users can create pages to share posters, talks or anything that might be relevant to the conference. In all, it could be a good playground to try out new ideas for online conferences. We could ask Nature to broadcast it in Nature island inside second life ? :)
Register, create your pages, share some thoughts. The topics are:
Career Development
Omics Session / Systems Biology
Scientific Communication 2.0
Konrad is one of the organizers.
Tags: open science, EMBL,conference
Tuesday, November 21, 2006
Connotea tag:evolution citation report
What is a scientific journal ? One possible definition could be - a content provider that filters and selects scientific content appropriate (of interest) to a particular group of people. Currently, journals select papers based on the decisions of a small group of people, maybe one or two editors supported by a few referees. The internet allows for alternative methods to select and filter content based potentially on the knowledge of a larger group of people. Eventually, these methods might one day replace the expansive editorial procedures now in place in most journals, but before that happens these approaches have to be evaluated. Also, even if we don't use these methods to replace current editorial procedures, they can be used to help us highlight the most interesting works published in certain fields.
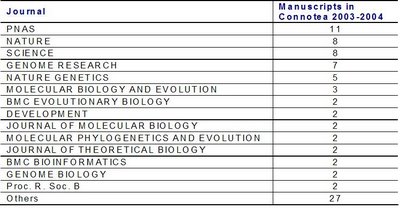
So, why not consider tags in social bookmarking services like Connotea as scientific journals ? Here is the journal Connotea tag:evolution. I took from Connotea yesterday (21/11/2006) all papers tagged with the tag "evolution" , that were published in 2003 or 2004 (85 papers). I used the web of science to get the number of citations of each of these papers (see figure below). This was unfortunately done one by one. I am thinking of scripting some tool to do it automatically but if someone knows a better way please let me know.
 As expected the most represented journals are some of the journals with higher visibility but still more than 50% of the manuscripts were tagged from more specialized journals.
As expected the most represented journals are some of the journals with higher visibility but still more than 50% of the manuscripts were tagged from more specialized journals.
 So how does the average number of citations per paper of this "new journal" compare with well established journals ?
So how does the average number of citations per paper of this "new journal" compare with well established journals ?
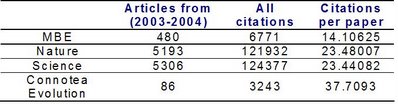 Although Connotea Evolution is low volume compared to other journals it does have a higher average citation per paper than journals such as Nature and Science. I did not separate potential non citable items from any of the groups so it should be a reasonable fair comparison.
Although Connotea Evolution is low volume compared to other journals it does have a higher average citation per paper than journals such as Nature and Science. I did not separate potential non citable items from any of the groups so it should be a reasonable fair comparison.
I think this suggests that we should evaluate potential mechanism to guide us to interesting scientific content. By itself, these evaluations might establish a form of reward for the community to come up with more sophisticated tools. It is important therefore to carefully pick the measurements. I used the citations per paper but others might be more adequate.
Any group of people can use the internet to re-group scientific content (specially if it is open access) into "journals" of potentially more value than those currently available.
What is a scientific journal ? One possible definition could be - a content provider that filters and selects scientific content appropriate (of interest) to a particular group of people. Currently, journals select papers based on the decisions of a small group of people, maybe one or two editors supported by a few referees. The internet allows for alternative methods to select and filter content based potentially on the knowledge of a larger group of people. Eventually, these methods might one day replace the expansive editorial procedures now in place in most journals, but before that happens these approaches have to be evaluated. Also, even if we don't use these methods to replace current editorial procedures, they can be used to help us highlight the most interesting works published in certain fields.
So, why not consider tags in social bookmarking services like Connotea as scientific journals ? Here is the journal Connotea tag:evolution. I took from Connotea yesterday (21/11/2006) all papers tagged with the tag "evolution" , that were published in 2003 or 2004 (85 papers). I used the web of science to get the number of citations of each of these papers (see figure below). This was unfortunately done one by one. I am thinking of scripting some tool to do it automatically but if someone knows a better way please let me know.
 As expected the most represented journals are some of the journals with higher visibility but still more than 50% of the manuscripts were tagged from more specialized journals.
As expected the most represented journals are some of the journals with higher visibility but still more than 50% of the manuscripts were tagged from more specialized journals. So how does the average number of citations per paper of this "new journal" compare with well established journals ?
So how does the average number of citations per paper of this "new journal" compare with well established journals ? Although Connotea Evolution is low volume compared to other journals it does have a higher average citation per paper than journals such as Nature and Science. I did not separate potential non citable items from any of the groups so it should be a reasonable fair comparison.
Although Connotea Evolution is low volume compared to other journals it does have a higher average citation per paper than journals such as Nature and Science. I did not separate potential non citable items from any of the groups so it should be a reasonable fair comparison.I think this suggests that we should evaluate potential mechanism to guide us to interesting scientific content. By itself, these evaluations might establish a form of reward for the community to come up with more sophisticated tools. It is important therefore to carefully pick the measurements. I used the citations per paper but others might be more adequate.
Any group of people can use the internet to re-group scientific content (specially if it is open access) into "journals" of potentially more value than those currently available.
Monday, November 20, 2006
Journals Proliferate
I only noticed today BMC also had a Systems Biology journal. There are no papers yet but the editorial board looks interesting enough. From the types of articles they expect I think they will take a lot of bioinformatics related manuscripts. This adds to two other Systems Biology journals that I am aware of: Molecular Systems Biology (also open access) and IET Systems Biology.
(via BioHacking) On a more creative note, here is the Journal of Visualized Experiments. It is a "journal" of recorded experiments that should help others learn protocols with the aid of videos. Currently the submissions are subjected only to editorial evaluation and are expected to get published in about 14 days. They plan to apply for listing in PubMed and other databases.
The videos are accompanied by a very short written explanation and are tagged for searching. There are no comments or RSS features that I could see.
Nature Methods and Nature Protocols should give this a try. By the way, Nature Protocols also publishes protocols in bioinformatics, and Nature Methods started a blog (Methagora)
With this continuing expansion of journals in all publishing houses aren't we quickly reaching a point when manuscripts will be the scarce resource ? I really hope someone develops nicer tools to suggest communications to read based on my interests. Is there a place for researchers whose job is just to associate and shuttle communications around ? Tagging communications as FOR_SMITH_J to show up in some reader with a comment: "solves your problem X".
I only noticed today BMC also had a Systems Biology journal. There are no papers yet but the editorial board looks interesting enough. From the types of articles they expect I think they will take a lot of bioinformatics related manuscripts. This adds to two other Systems Biology journals that I am aware of: Molecular Systems Biology (also open access) and IET Systems Biology.
(via BioHacking) On a more creative note, here is the Journal of Visualized Experiments. It is a "journal" of recorded experiments that should help others learn protocols with the aid of videos. Currently the submissions are subjected only to editorial evaluation and are expected to get published in about 14 days. They plan to apply for listing in PubMed and other databases.
The videos are accompanied by a very short written explanation and are tagged for searching. There are no comments or RSS features that I could see.
Nature Methods and Nature Protocols should give this a try. By the way, Nature Protocols also publishes protocols in bioinformatics, and Nature Methods started a blog (Methagora)
With this continuing expansion of journals in all publishing houses aren't we quickly reaching a point when manuscripts will be the scarce resource ? I really hope someone develops nicer tools to suggest communications to read based on my interests. Is there a place for researchers whose job is just to associate and shuttle communications around ? Tagging communications as FOR_SMITH_J to show up in some reader with a comment: "solves your problem X".
Chris Surridge explains PLoS ONE
As part of the OpenWetWare's Seminar Series on Open Science, here is Chris Surridge explaining PLoS ONE. Some random things I remember:
- No-one reads full table of contents anymore so why not create a journal with broad scope ? (I do read full table of contents of many journals, it's the first thing I do in the morning)
- They are aiming at a very high volume (hundreds of manuscripts)
- The journal will probably have portals for subject areas
- Anyone is free to reuse the open access content, so anyone could in theory be an editor by reusing open access content and focusing it for a particular target audience.
- They might also pool in papers from other open access journals
- There will be probably a karma system to rate the contributions
- The system of having different versions of the same manuscript will not be on the first version of the journal.
- The journal might be up on the 29th of this month (not sure yet)
- Chris Surridge moves around too much :)
Tags: plos,plosone,open science
Tuesday, November 14, 2006
iTOL - Interactive Tree Of Life
The Bork group published recently a revised tree of life in Science. You can have a better look at the tree using this nice interactive tree viewer that they put up on the web (publication).

You can zoom in and out, re-root, swap branches, and export the trees. You can also upload your own trees and annotations to have them drawn by the viewer. Hovering over the species brings up a box with some information. Did you ever wonder who donated DNA for the human sequencing project ? The Bork group has found out for us:

The Bork group published recently a revised tree of life in Science. You can have a better look at the tree using this nice interactive tree viewer that they put up on the web (publication).

You can zoom in and out, re-root, swap branches, and export the trees. You can also upload your own trees and annotations to have them drawn by the viewer. Hovering over the species brings up a box with some information. Did you ever wonder who donated DNA for the human sequencing project ? The Bork group has found out for us:

Monday, November 13, 2006
PLoS redesigns, PLoS ONE soon
Go have a look at the PLoS websites, they have been redesigned. I like the look but the only notable changes are a box "From the Blogosphere" that currently links to the Open Access News blog (I wonder why:) and a link to "Readers Respond" on the left, that should put more emphasis on user participation. The PLoS Medicine site has some notable differences. First they include the PLoS Medicine blog on the home page and the "From the Blogosphere" links to a Guardian article. I guess that these are customizable and left to the editors to use to point out interesting things related to the journal or field. Now ... when will PLoS Comp Bio start a blog ?
Still no journal is taking in blog comments. It would be easy to use Postgenomic's index or trackbacks to let readers comments papers trough their blogs.
There is also an editorial in PLoS Biology about PLoS ONE: "ONE for All: The Next Step for PLoS"
The only new thing I got from the editorial was the concept of portals withing PLoS ONE. I would say it sounds a bit like the Nature Gateways, an area for the aggregation of papers and other resources related to a particular field or project. Sounds like a good idea. Again, they mention that at the start it will look like any other journal and that they will build on it in time, so I don't expect much in the launch day.
Go have a look at the PLoS websites, they have been redesigned. I like the look but the only notable changes are a box "From the Blogosphere" that currently links to the Open Access News blog (I wonder why:) and a link to "Readers Respond" on the left, that should put more emphasis on user participation. The PLoS Medicine site has some notable differences. First they include the PLoS Medicine blog on the home page and the "From the Blogosphere" links to a Guardian article. I guess that these are customizable and left to the editors to use to point out interesting things related to the journal or field. Now ... when will PLoS Comp Bio start a blog ?
Still no journal is taking in blog comments. It would be easy to use Postgenomic's index or trackbacks to let readers comments papers trough their blogs.
There is also an editorial in PLoS Biology about PLoS ONE: "ONE for All: The Next Step for PLoS"
The only new thing I got from the editorial was the concept of portals withing PLoS ONE. I would say it sounds a bit like the Nature Gateways, an area for the aggregation of papers and other resources related to a particular field or project. Sounds like a good idea. Again, they mention that at the start it will look like any other journal and that they will build on it in time, so I don't expect much in the launch day.
Tags: plos,plos one,open access
Subscribe to:
Comments (Atom)