Specificity and Evolvability in Protein Interaction NetworksI finally have the opportunity to blog about some of what I have worked on during last year. It has been published in PLoS Computational Biology and is freely
available here in the early online release format (still in the original ugly format :). One way to use our blogs may be to add some depth to the papers that we published. Something like the extras we get when we buy the DVD of a movie ;)
Main conclusions- Protein interactions can change at a fast rate of 1E-5 interactions per protein pair per million years
- Binding specificity is a strong determining of binding specificity with more promiscuous binding proteins having a higher rate of change of interactions.
- Human proteins involved in immune response, transport and establishment of localization, show signs of positive selection for change of interactions.
The making ofWe
had been using comparative genomics to search for conserved putative protein binding sites. These very conserved putative target sites were very likely to be experimentally known target sites but many other known binding sites seamed not to be so conserved. This was what got us started thinking about the evolution of these protein interactions and what might determine the rate at which interactions are gained and lost during evolution. The analysis was mostly inspired on the nice work of
Andreas Wagner that
first proposed a rate for the addition of new interactions in
S. cerevisiae. We have tried to build on this by analyzing different species and determining also what protein properties might determine the rate at which interactions are gained and lost in evolution.
More than nodes and edgesOne of the main conclusions from this work was that binding specificity also determines the rate of change of interactions during evolution. More promiscuous binding proteins not only have many binding partners but they also tend to change partners faster during evolution. To establish a proxy for binding specificity we have used structural information from the
iPFAM database. In essence we have considered that protein domains that have been seen in contact with many different other domains would be more promiscuous. In general we observed that proteins containing these promiscuous domains had a high rate of change of interactions (see figure below).
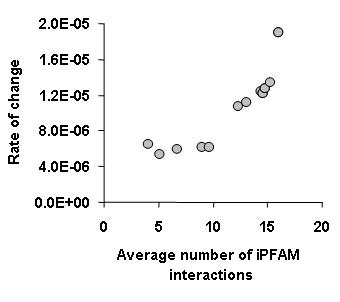
This highlights something that I
had stressed before, that it is important to consider protein interaction networks as more than nodes and edges. Another recent paper has also shown that it is possible and useful to use the accumulating structural information available in the PDB to obtain a more accurate representation of protein interaction networks. Philip M. Kim and co workers from the Gerstein lab (
blog,
webpage) published a
study in Science were they have used also the iPFAM database to curate all the
S. cerevisie interactions and to discriminate between interactions that use the same or different binding interfaces. With this information the authors distinguish between hubs that tend to interact with their partners mostly trough one interface or trough many interfaces. They have shown that the multi interface hubs are more restricted in evolution and more likely to be essential than single interface hubs. They have a
website with presentations and additional data for this paper.
In the pipelineTo be submitted soon (hopefully), are some collaborations on how to use structural information to predict protein-protein binding specificity. With these collaborations I finish my thesis (still waiting for the defense). During the next couple of months I am off to search for a lab to work as a postdoc.