The value of a reader in an author pays publishing model
In many respects the changes in online communication and collaboration have been the leading edge of what latter is tested by the scientific community. I regard sites like Digg, del.icio.us, blogger and other related sites as experiments from witch we can learn about using the internet to make scientific communication and collaboration more efficient. In that context I think there might be an interesting analogy between a study (PDF version), pointed by Nick Carr discussing the value of a free costumer.
In this study the authors created a model to analyze the "profitability of costumers in a networked setting". One example of this type of setting are the auction houses were two distinct users (buyers and sellers) exist. Buyers do not pay anything to the auction houses but provide an obvious value that is, as they say, difficult to quantify. In their analysis they estimated that in this type of network setting the value of buyer is actually higher than the value of the seller (the one that actually pays to use the service).
How might this relate to scientific publishing? In the current model of a journal like PLoS or similar journals there are two very obvious "costumers", the author (that pays to have the article distributed) and the readers, that pay nothing to get access to the journal. The other main publishing model is the opposite, the authors pay nothing (or much less at least) and the readers have to pay to access the journal. It might be that in the different models the publishers might have to direct their efforts differently. In a journal like PLoS ONE were the quality of the service might actually improve with the participation of the readers (annotations/discussions) I would think that the value of the reader is likely much higher than the paying costumer (authors). It would interesting to read a similar study directed at the economics of scientific publishing.
Tuesday, March 06, 2007
OpenWetWare:Reviews
Jason Kelly opened up a page in OpenWetWare for discussion of wiki reviews. Writing a review on the progress of a field could be the most obvious use of collaborative efforts. The reviews could be continually updated and periodic versions could be frozen and submitted to a more conventional repository.
Jason suggests that one way to kick start the process would be to wikify a review published already with an open license and let people update it. OpenWetWare has now over 2000 registered users and continues to grow as probably the best example of online collaborations in science.
If you have ideas about how to implement wiki reviews, or simple want to start writing one, head over there and give it a try.
Jason Kelly opened up a page in OpenWetWare for discussion of wiki reviews. Writing a review on the progress of a field could be the most obvious use of collaborative efforts. The reviews could be continually updated and periodic versions could be frozen and submitted to a more conventional repository.
Jason suggests that one way to kick start the process would be to wikify a review published already with an open license and let people update it. OpenWetWare has now over 2000 registered users and continues to grow as probably the best example of online collaborations in science.
If you have ideas about how to implement wiki reviews, or simple want to start writing one, head over there and give it a try.
Sunday, March 04, 2007
Blogroll update
I finally got around to setting up the blogroll after updating the blog to the new blogger version. I have put on the right side most of the things that I am enjoying reading at the moment, separated in four sections:
Bioinformatics
Biotech & Drug Discovery
Evolution & Genomics
Publishing & General Science
One novelty in the list is the BioMed Central blog. They are now the third publishing house with some sort of official blog. Nature was the first and at least Nascent, Nautilus and Peer-to-peer provide mostly useful information and a way to interact with Nature services. Some other Nature blogs are not as interesting, in part because of a disturbing habit of using blog posts has a mirror for the table of contents of the journal.
I finally got around to setting up the blogroll after updating the blog to the new blogger version. I have put on the right side most of the things that I am enjoying reading at the moment, separated in four sections:
Bioinformatics
Biotech & Drug Discovery
Evolution & Genomics
Publishing & General Science
One novelty in the list is the BioMed Central blog. They are now the third publishing house with some sort of official blog. Nature was the first and at least Nascent, Nautilus and Peer-to-peer provide mostly useful information and a way to interact with Nature services. Some other Nature blogs are not as interesting, in part because of a disturbing habit of using blog posts has a mirror for the table of contents of the journal.
Thursday, March 01, 2007
Bio::Blogs #8
Editorial musings

Welcome to the eight edition of the bioinformatics blog journal Bio::Blogs. The archive from previous months can be found at bioblogs.wordpress.com. When this carnival was started, more than eight months ago, it had the primary objective to serve as sort of display for some of the best bioinformatics blog posts on the web and to create incentives for other people to promote their blogs and join in the conversation.
Looking back at the past Bio::Blogs editions I would like to think that we have manage to come with with many interesting posts about bioinformatic conferences, tools and useful computational tips like the DNA analysis series by Sandra Porter (I,II,III,IV,V,VI). Bio::Blogs has also been use to promote tools that have been published in blogs like the Genetic Programming Applet from Pierre and research like the correlation between protein-interaction likelihood and protein age (I and II) that I worked on.
Not everything went as I would hope. The submissions to Bio::Blogs have not picked up as I would expect. Some of this can be explained by poor promotion of my part but it is also due to the small size of the bioinformatics blogging community. In any case I think it is worth maintaining Bio::Blogs up and running for some more time before thinking about stopping this experiment.
In this edition a PDF version of all the posts has been created for anyone interested in downloading, printing and reading some fine posts over coffee or tea. Leave comments or send an email (bioblogs at gmail.com) with your thoughts/ideas for the continuation of this blog journal.I think this printed version also gives a more concrete impression of the potential of blogging for scientific communication.
News and Views
GrrlScientist submitted a report on a recent Science paper describing how moths use their antennae to help them stabilize their flight. Apart from some nasty experiments were they authors removed and glued parts of the antenna the work features some interesting neurophysiology analysis used to characterize the sensitivity of the antennae.
From my blog I picked an entry on network reconstruction. I think the increasing amounts of omics data should be better explored than it currently is and network reconstruction methods are a very good way of achieving this. In this paper, Jason Ernst and colleagues used expression data to reconstruct dynamic transcription regulatory interactions. I will try to continue blogging about the topic in future posts.
Comentaries
PLoS ONE was launched about two months ago and it has produced so far an amazing stream of publications. However the initially proposed goal of generating discussions online to promote post-publication reviews as been lagging. Stew and Alf wrote two commentaries (summited by Greg) regarding the progress of PLoS ONE. They both discuss the current lack of infrastructures to stimulate the online discussions at the PLoS ONE site. Stew goes even further by providing to anyone interested a nice Gresemonkey script to add blog comments to the PLoS ONE papers. I hope Chris Surridge and the rest of the PLoS ONE team start deploying soon some of the tools that they have talked about in their blog. They need to make ONE feel like home, a place were a community of people can discuss their papers.
From Deepak we have a post dedicated to what he dubs EcoInformatics. The importance of using computational methods to analyze ecological changes from the molecules to the ecosystems. The problems range from data access to data management and analysis. The complexity and the different scales of organization (i.e. molecules, environment, ecosystems, disease) make this a very promising field for computational biologists.
From ecological changes we move on to human evolution. Phil tries to introduce the possibility of humanity using technology to improve itself. Having a strong interest myself in synthetic biology and man-machine interfaces I would say that we are still far away from having such control. It is nevertheless useful to discuss the implications of emerging technologies to better prepare for the possible changes.
Reviews and tips
I start this section with a post from a bran-new bioinformatics blog called Bioinformatics Zen. Michael Barton submitted his post on useful tips to get organized as a dry lab scientist. I agree with most of his suggestions. I have try to use slightly fancier methods of organizing my work using project managing tools but I end up returning to a more straightforward folder based approach as well.
Neil Saunders sent in a nice tutorial on building AJAX pages for bioinformatics. It is a very well explained introduction with annotated code. If you were ever interested in learning the basics of AJAX but never invested time in it, here is a good chance to try it.
Editorial musings

Welcome to the eight edition of the bioinformatics blog journal Bio::Blogs. The archive from previous months can be found at bioblogs.wordpress.com. When this carnival was started, more than eight months ago, it had the primary objective to serve as sort of display for some of the best bioinformatics blog posts on the web and to create incentives for other people to promote their blogs and join in the conversation.
Looking back at the past Bio::Blogs editions I would like to think that we have manage to come with with many interesting posts about bioinformatic conferences, tools and useful computational tips like the DNA analysis series by Sandra Porter (I,II,III,IV,V,VI). Bio::Blogs has also been use to promote tools that have been published in blogs like the Genetic Programming Applet from Pierre and research like the correlation between protein-interaction likelihood and protein age (I and II) that I worked on.
Not everything went as I would hope. The submissions to Bio::Blogs have not picked up as I would expect. Some of this can be explained by poor promotion of my part but it is also due to the small size of the bioinformatics blogging community. In any case I think it is worth maintaining Bio::Blogs up and running for some more time before thinking about stopping this experiment.
In this edition a PDF version of all the posts has been created for anyone interested in downloading, printing and reading some fine posts over coffee or tea. Leave comments or send an email (bioblogs at gmail.com) with your thoughts/ideas for the continuation of this blog journal.I think this printed version also gives a more concrete impression of the potential of blogging for scientific communication.
News and Views
GrrlScientist submitted a report on a recent Science paper describing how moths use their antennae to help them stabilize their flight. Apart from some nasty experiments were they authors removed and glued parts of the antenna the work features some interesting neurophysiology analysis used to characterize the sensitivity of the antennae.
From my blog I picked an entry on network reconstruction. I think the increasing amounts of omics data should be better explored than it currently is and network reconstruction methods are a very good way of achieving this. In this paper, Jason Ernst and colleagues used expression data to reconstruct dynamic transcription regulatory interactions. I will try to continue blogging about the topic in future posts.
Comentaries
PLoS ONE was launched about two months ago and it has produced so far an amazing stream of publications. However the initially proposed goal of generating discussions online to promote post-publication reviews as been lagging. Stew and Alf wrote two commentaries (summited by Greg) regarding the progress of PLoS ONE. They both discuss the current lack of infrastructures to stimulate the online discussions at the PLoS ONE site. Stew goes even further by providing to anyone interested a nice Gresemonkey script to add blog comments to the PLoS ONE papers. I hope Chris Surridge and the rest of the PLoS ONE team start deploying soon some of the tools that they have talked about in their blog. They need to make ONE feel like home, a place were a community of people can discuss their papers.
From Deepak we have a post dedicated to what he dubs EcoInformatics. The importance of using computational methods to analyze ecological changes from the molecules to the ecosystems. The problems range from data access to data management and analysis. The complexity and the different scales of organization (i.e. molecules, environment, ecosystems, disease) make this a very promising field for computational biologists.
From ecological changes we move on to human evolution. Phil tries to introduce the possibility of humanity using technology to improve itself. Having a strong interest myself in synthetic biology and man-machine interfaces I would say that we are still far away from having such control. It is nevertheless useful to discuss the implications of emerging technologies to better prepare for the possible changes.
Reviews and tips
I start this section with a post from a bran-new bioinformatics blog called Bioinformatics Zen. Michael Barton submitted his post on useful tips to get organized as a dry lab scientist. I agree with most of his suggestions. I have try to use slightly fancier methods of organizing my work using project managing tools but I end up returning to a more straightforward folder based approach as well.
Neil Saunders sent in a nice tutorial on building AJAX pages for bioinformatics. It is a very well explained introduction with annotated code. If you were ever interested in learning the basics of AJAX but never invested time in it, here is a good chance to try it.
Craig Venter in Colbert Report
(via Drew Endy in SynBio discuss list) Here is mister Craig Venter in Colbert Report promoting synthetic biology and the personal genome (in a funny way).
Let's hope that Synthetic Biology does not get over hyped. The public might start reacting negatively to these technologies if they grow too fast or if they don't deliver what they promise.
(via Drew Endy in SynBio discuss list) Here is mister Craig Venter in Colbert Report promoting synthetic biology and the personal genome (in a funny way).
Let's hope that Synthetic Biology does not get over hyped. The public might start reacting negatively to these technologies if they grow too fast or if they don't deliver what they promise.
Wednesday, February 28, 2007
Bio::Blogs #8
The eighth edition of Bio::Blogs is coming up soon. It is going to be published here on Public Rambling tomorrow night. I almost forgot that this month is shorter than usual :). There are currently 3 submissions sent in. Pick something from your blogs that is bioinformatic related and sent it in (bioblogs at gmail) or leave the link in the comments. Please also let me know if you mind that I create a PDF file including your submission for offline reading. If you don;t have a blog start one and let me know or just send in a link to something that you particularly liked.
The eighth edition of Bio::Blogs is coming up soon. It is going to be published here on Public Rambling tomorrow night. I almost forgot that this month is shorter than usual :). There are currently 3 submissions sent in. Pick something from your blogs that is bioinformatic related and sent it in (bioblogs at gmail) or leave the link in the comments. Please also let me know if you mind that I create a PDF file including your submission for offline reading. If you don;t have a blog start one and let me know or just send in a link to something that you particularly liked.
Tuesday, February 27, 2007
The future impact of genome synthesis
The synthesis blog pointed to a detailed report discussing the economical importance of impending advances in biological engineering. The study, supported by DOE, DuPont Corporation and The Berkley Nanosciences Nanoengineering Institute tries to cover the main driving forces for biotechnology innovation, it's possible future applications and economical impact. The last chapter is dedicated to envisioning future scenarios for synthetic biology based on different assumptions about important factors that could determine the progress of this technology.
While the scenarios described in the end of the report might be useful to track the speed and mode of evolution of this emerging technology, the most relevant section for life scientists is arguably the one discussing possible applications of genome synthesis.
There are three main applications listed:
Chemicals: Engineering new production pathways and creating new products
Energy: Opening new biological routes for energy transformation
Synthetic Vaccines: Opportunities for rapid-response biosecurity
 The best examples of synthetic biology research have consisted up to now mostly of simple toy examples. Usually simple circuits are created and studied to detail but few have obvious immediate practical applications. Are we currently at the inflection point, were synthetic biology research will produce more practical applications or is the complexity of living systems still too large a barrier ?
The best examples of synthetic biology research have consisted up to now mostly of simple toy examples. Usually simple circuits are created and studied to detail but few have obvious immediate practical applications. Are we currently at the inflection point, were synthetic biology research will produce more practical applications or is the complexity of living systems still too large a barrier ?
One example of the use of synthetic biology in chemical production is the work of Dae-Kyun Ro and colleagues in the Keasling lab (free PDF). They re-engineered S. cerevisiae to produce artemisinic acid, a precursor of the malaria drug Artemisinin.
Keasling is also one of the researchers involved in the Helios project. An effort directed at developing technology for solar fuel generation (in the form of biofuel). The project is also headed by Nobel prize laureate Steve Chu that explains the project in this video presentation.
The synthesis blog pointed to a detailed report discussing the economical importance of impending advances in biological engineering. The study, supported by DOE, DuPont Corporation and The Berkley Nanosciences Nanoengineering Institute tries to cover the main driving forces for biotechnology innovation, it's possible future applications and economical impact. The last chapter is dedicated to envisioning future scenarios for synthetic biology based on different assumptions about important factors that could determine the progress of this technology.
While the scenarios described in the end of the report might be useful to track the speed and mode of evolution of this emerging technology, the most relevant section for life scientists is arguably the one discussing possible applications of genome synthesis.
There are three main applications listed:
Chemicals: Engineering new production pathways and creating new products
Energy: Opening new biological routes for energy transformation
Synthetic Vaccines: Opportunities for rapid-response biosecurity
 The best examples of synthetic biology research have consisted up to now mostly of simple toy examples. Usually simple circuits are created and studied to detail but few have obvious immediate practical applications. Are we currently at the inflection point, were synthetic biology research will produce more practical applications or is the complexity of living systems still too large a barrier ?
The best examples of synthetic biology research have consisted up to now mostly of simple toy examples. Usually simple circuits are created and studied to detail but few have obvious immediate practical applications. Are we currently at the inflection point, were synthetic biology research will produce more practical applications or is the complexity of living systems still too large a barrier ?One example of the use of synthetic biology in chemical production is the work of Dae-Kyun Ro and colleagues in the Keasling lab (free PDF). They re-engineered S. cerevisiae to produce artemisinic acid, a precursor of the malaria drug Artemisinin.
Keasling is also one of the researchers involved in the Helios project. An effort directed at developing technology for solar fuel generation (in the form of biofuel). The project is also headed by Nobel prize laureate Steve Chu that explains the project in this video presentation.
Friday, February 23, 2007
Traveling around (Boston, San Francisco)
I have been traveling during the past week. I have been in Boston and I am now in San Francisco (labs: Marc Vidal, Wendel Lim, Adam Arkin).It would be interesting to be able to talk about some of the nice projects I have heard about but I guess it is really not up to me to make this public. Some of it is on their webpages. That leaves very little to say about the trip in respect to science. So instead, here is a picture I took in San Francisco :)

I am looking for a place to start a postdoc after the summer time. Even if I don't move to the states it is very unlikely that I will stay in Germany. This will be my 3rd country and 7th city. It is funny that so many of the grants that are currently available in Europe for postdocs are incentives to increase mobility. Isn't it time to also create some incentives to settling down ? I am 28 and this will be my first postdoc but eventually I will get tired of moving around. I hope by then it will be easier to stay.
I have been traveling during the past week. I have been in Boston and I am now in San Francisco (labs: Marc Vidal, Wendel Lim, Adam Arkin).It would be interesting to be able to talk about some of the nice projects I have heard about but I guess it is really not up to me to make this public. Some of it is on their webpages. That leaves very little to say about the trip in respect to science. So instead, here is a picture I took in San Francisco :)

I am looking for a place to start a postdoc after the summer time. Even if I don't move to the states it is very unlikely that I will stay in Germany. This will be my 3rd country and 7th city. It is funny that so many of the grants that are currently available in Europe for postdocs are incentives to increase mobility. Isn't it time to also create some incentives to settling down ? I am 28 and this will be my first postdoc but eventually I will get tired of moving around. I hope by then it will be easier to stay.
Wednesday, February 07, 2007
in sillico network reconstruction (using expression data)
In my last post I commented on a paper that tried to find the best mathematical model for a cellular pathway. In that paper they used information on known and predicted protein interactions. This time I want to mention a paper, published in Nature Mol. Systems Biology, that attempts to reconstruct gene regulatory networks from gene expression data and Chip-chip data.
The authors were interested in determining how/when transcription factors regulate their target genes over time. One novelty introduced in this work was the focus on bifurcation events in gene expression. They tried to look for cases where a groups of genes clearly bifurcated into two groups at a particular time point. Combining these patterns of bifurcation with experimental binding data for transcription factors they tried to predict what transcription factors regulate these group of genes. There is a simple example shown in figure 1, reproduced below.

In this toy example there is a bifurcation event at 1 h and another at the 2h time point. All of the genes are assigned to a gene expression path. In this case, the red genes are those that are very likely to show a down regulation in between the 1st and 2nd hour and stay at the same level of expression from then on. Once the genes have been assigned it is possible to search for transcription factors that are significantly associate to each gene expression path. For example in this case, TF A is strongly associated to the pink trajectory. This means that many of the genes in the pink group have a known binding site for TF A in their promoter region.
To test their approach, the authors studied the amino-acid starvation in S. cerevisiae. In figure 2 they summarize the reconstructed dynamic map. The result is the association of TFs to groups of genes and the changes in expression of these genes over time during amino acid starvation.

One interesting finding from this map was that Ino4 activates a group of genes related to lipid metabolism starting at the 2h time point. Since Ino4 binding sites had only been profiled by Chip-chip in YPD media and not in a.a. starvation, this is a novel result obtained using their method.
To further test the significance of their observation they performed Chip-chip assays of Ino4 in amino acid starvation. They confirmed that Ino4 binds many more promoters during amino acid starvation as compared to synthetic complete glucose media. Out of 207 genes bound by Ino4 (specifically during AA starvation) 34 were also among the genes assigned to the Ino4 gene path obtained from their approach.
This results confirmed the usefulness of this computational approach to reconstruct gene regulatory networks from gene expression data and TF binding site information.
The authors then go on to study the regulation of other conditions.
For anyone curious enough about the method, this was done using Hidden Markov Models (see here for available primer on HMMs).
In my last post I commented on a paper that tried to find the best mathematical model for a cellular pathway. In that paper they used information on known and predicted protein interactions. This time I want to mention a paper, published in Nature Mol. Systems Biology, that attempts to reconstruct gene regulatory networks from gene expression data and Chip-chip data.
The authors were interested in determining how/when transcription factors regulate their target genes over time. One novelty introduced in this work was the focus on bifurcation events in gene expression. They tried to look for cases where a groups of genes clearly bifurcated into two groups at a particular time point. Combining these patterns of bifurcation with experimental binding data for transcription factors they tried to predict what transcription factors regulate these group of genes. There is a simple example shown in figure 1, reproduced below.

In this toy example there is a bifurcation event at 1 h and another at the 2h time point. All of the genes are assigned to a gene expression path. In this case, the red genes are those that are very likely to show a down regulation in between the 1st and 2nd hour and stay at the same level of expression from then on. Once the genes have been assigned it is possible to search for transcription factors that are significantly associate to each gene expression path. For example in this case, TF A is strongly associated to the pink trajectory. This means that many of the genes in the pink group have a known binding site for TF A in their promoter region.
To test their approach, the authors studied the amino-acid starvation in S. cerevisiae. In figure 2 they summarize the reconstructed dynamic map. The result is the association of TFs to groups of genes and the changes in expression of these genes over time during amino acid starvation.

One interesting finding from this map was that Ino4 activates a group of genes related to lipid metabolism starting at the 2h time point. Since Ino4 binding sites had only been profiled by Chip-chip in YPD media and not in a.a. starvation, this is a novel result obtained using their method.
To further test the significance of their observation they performed Chip-chip assays of Ino4 in amino acid starvation. They confirmed that Ino4 binds many more promoters during amino acid starvation as compared to synthetic complete glucose media. Out of 207 genes bound by Ino4 (specifically during AA starvation) 34 were also among the genes assigned to the Ino4 gene path obtained from their approach.
This results confirmed the usefulness of this computational approach to reconstruct gene regulatory networks from gene expression data and TF binding site information.
The authors then go on to study the regulation of other conditions.
For anyone curious enough about the method, this was done using Hidden Markov Models (see here for available primer on HMMs).
Tuesday, February 06, 2007
In silico network reconstruction
It is day one of Just Science week and I want to tell you about a recent paper that was published in BMC Systems Biology by Rui Alves and Albert Sorribas. It is about a general approach to integrate information to come up with models for cellular pathways. What does this mean and why is this important ?
Increasingly the scientific knowledge is being stored in databases (literature, protein structures, gene expression, protein-protein interactions, protein-DNA interactions, etc). The general idea behind the work described is that we should be able to use the accumulated information about cellular pathways to extract models of how the cell's components interact to preform their functions. By models I mean a formal representation that can tell us how the components' concentrations and activities change with time.
There are several works already dealing with this problem of trying to reconstruct cellular networks from large data sources but I found this article particularly interesting because it uses so many of these methods.
To give you an idea I reproduce below figure 4 of the paper with a diagram of the method (click to zoom in):

The authors have pulled in experimentally known interactions and combined them with putative interactions obtained from docking and phylogenetic based predictions. These predicted networks are then converted to several possible mathematical models that are examined under different parameter conditions and compared with known experimental values.
This method should be particularly suited for a case when some of the genes in the pathway are known and there are experimental measured outputs for the pathway that can be compared with the predictions from the putative pathway models.
Ideally this whole procedure would be fully converted into an automatic pipeline that could be used by people that are not so familiar with the tools.
I will try to stick with the same theme during the week, hopefully covering different methods to achieve the same thing.
It is day one of Just Science week and I want to tell you about a recent paper that was published in BMC Systems Biology by Rui Alves and Albert Sorribas. It is about a general approach to integrate information to come up with models for cellular pathways. What does this mean and why is this important ?
Increasingly the scientific knowledge is being stored in databases (literature, protein structures, gene expression, protein-protein interactions, protein-DNA interactions, etc). The general idea behind the work described is that we should be able to use the accumulated information about cellular pathways to extract models of how the cell's components interact to preform their functions. By models I mean a formal representation that can tell us how the components' concentrations and activities change with time.
There are several works already dealing with this problem of trying to reconstruct cellular networks from large data sources but I found this article particularly interesting because it uses so many of these methods.
To give you an idea I reproduce below figure 4 of the paper with a diagram of the method (click to zoom in):

The authors have pulled in experimentally known interactions and combined them with putative interactions obtained from docking and phylogenetic based predictions. These predicted networks are then converted to several possible mathematical models that are examined under different parameter conditions and compared with known experimental values.
This method should be particularly suited for a case when some of the genes in the pathway are known and there are experimental measured outputs for the pathway that can be compared with the predictions from the putative pathway models.
Ideally this whole procedure would be fully converted into an automatic pipeline that could be used by people that are not so familiar with the tools.
I will try to stick with the same theme during the week, hopefully covering different methods to achieve the same thing.
Sunday, February 04, 2007
Publishing greasemonkey scripts (update)
A while ago I asked if greasemonkey scripts should be published in peer reviewed journals or if blogs could be a more suitable way of distributing these tools. The blog post was triggered by the publication of iHOPerator, mentioned also by Deepak.
I would like to thank one of the authors, Benjamin Good, and a BMC editor, Matt Hodgkinson, for taking the time to post their opinion in the comments. In summary they both argue that this publication helps raise awareness to greasemonkey and related technologies.
For me this exchange in the comments exemplifies the usefulness of the web for discussing science. The comments on this paper are aggregated in this Postgenomic entry and anyone could, in principle, participate no matter where they are.
This also reminded me of a discussion I had with someone here at EMBL recently. If web based discussions like this take off then authors might have a higher work load in trying to keep up with what is being said about their works. If a misinterpretation occurs it has a higher potential for spreading online. On the other hand, these sorts of web discussion help to level the playing field for manuscripts. In the near future it might not matter so much were the paper is published but if attracted the attention of the people in the field.
A while ago I asked if greasemonkey scripts should be published in peer reviewed journals or if blogs could be a more suitable way of distributing these tools. The blog post was triggered by the publication of iHOPerator, mentioned also by Deepak.
I would like to thank one of the authors, Benjamin Good, and a BMC editor, Matt Hodgkinson, for taking the time to post their opinion in the comments. In summary they both argue that this publication helps raise awareness to greasemonkey and related technologies.
For me this exchange in the comments exemplifies the usefulness of the web for discussing science. The comments on this paper are aggregated in this Postgenomic entry and anyone could, in principle, participate no matter where they are.
This also reminded me of a discussion I had with someone here at EMBL recently. If web based discussions like this take off then authors might have a higher work load in trying to keep up with what is being said about their works. If a misinterpretation occurs it has a higher potential for spreading online. On the other hand, these sorts of web discussion help to level the playing field for manuscripts. In the near future it might not matter so much were the paper is published but if attracted the attention of the people in the field.
Friday, February 02, 2007
Just Science
(Via RPM, Razib, Chris, Arunn) Next week is Just Science week. I will try to review recent papers on cellular networks, systems/synthetic biology and evolution that I found interesting.
(Via RPM, Razib, Chris, Arunn) Next week is Just Science week. I will try to review recent papers on cellular networks, systems/synthetic biology and evolution that I found interesting.
Bio::Blogs #7
The February edition of Bio::Blogs was just published in BioHacking. Thanks to Paras for editing it. He highlighted some blogs related to synthetic biology and some of the recent bioinformatics posts from Neil, Sandra and Pierre.
The 8th edition will be coming back here. The participation has been generally going down so Bio::Blogs might, in the near future, morph to something else.
Just to give it a little twist I thought that it could be interesting to add a PDF version of Bio::Blogs (with the permission of the authors of course). So, for the next month entries I will be asking if the authors concede that the blog posts be compiled into a printable document.
Entries can be submitted until the end of February to bioblogs at gmail.
The February edition of Bio::Blogs was just published in BioHacking. Thanks to Paras for editing it. He highlighted some blogs related to synthetic biology and some of the recent bioinformatics posts from Neil, Sandra and Pierre.
The 8th edition will be coming back here. The participation has been generally going down so Bio::Blogs might, in the near future, morph to something else.
Just to give it a little twist I thought that it could be interesting to add a PDF version of Bio::Blogs (with the permission of the authors of course). So, for the next month entries I will be asking if the authors concede that the blog posts be compiled into a printable document.
Entries can be submitted until the end of February to bioblogs at gmail.
Monday, January 29, 2007
Bio::Blogs #7 final call

As Deepak mentioned in his blog, the February edition of Bio::Blogs (the bioinformatics blog journal) is due soon. It will be up on the 2nd of February at BioHacking, so get your blogging pens working. It has been 2 months since the last call. Go have a look at the past two months, pick up one of your posts or a post you liked, related to bioinformatics and send it to the usual bioblogs at gmail.
The 8th edition will be back here on this blog. If anyone is interested in hosting future editions let me know in the comments or by email and I can make a list (including previous editors :).
Let's see how many posts highlighted in Bio::Blogs make it to the next years Science Blogging Anthology ;).

As Deepak mentioned in his blog, the February edition of Bio::Blogs (the bioinformatics blog journal) is due soon. It will be up on the 2nd of February at BioHacking, so get your blogging pens working. It has been 2 months since the last call. Go have a look at the past two months, pick up one of your posts or a post you liked, related to bioinformatics and send it to the usual bioblogs at gmail.
The 8th edition will be back here on this blog. If anyone is interested in hosting future editions let me know in the comments or by email and I can make a list (including previous editors :).
Let's see how many posts highlighted in Bio::Blogs make it to the next years Science Blogging Anthology ;).
Friday, January 26, 2007
Not so silent mutations
DNA mutations that do not change the coding amino-acid are many times referred to as "silent mutations", or synonymous mutations, because it is less likely that they will result in a change in function. Synonymous mutations are often considered to be evolutionary neutral and the ratio of non-synonymous substitutions (Ka) to synonymous substitutions (Ks) is used to study sequence evolution. It can be used for example to search for DNA regions targeted by selection (see review and a practical application).
In the last issue of Science Kimchi-Sarfaty and colleagues found a synonymous mutation in a transport protein that has an effect on the protein function. They have shown, at least in cell-lines, that the mutation does not affect mRNA levels nor the produced protein sequence. Finally the authors showed that the mutation might change the protein's conformation by comparing the sensibility of wild type and mutated sequence to trypsin digestion.
The authors speculate that the usage of that particular codon, even if not affecting the coding region, might change the translation rate and folding of the protein. It had already been shown in E. coli that synonymous mutations can affect the in vivo folding of a protein. Here the authors have shown a case where a silent mutation can change the substrate specificity of a transporter.
Because of these codon preferences it is important to adjust for codon selection pressures when studying synonymous substitutions. The codon preferences are usually considered to be due to differences in the pool of the cognate tRNA but other studies have shown that codon bias might arise also by codon context. In E. coli, codon pair preferences, were observed to affect their in vivo translation. Also, these codon pair preferences are species specific and are, at least in part, influenced by nucleotide positions within A-site tRNA sequences.
Hypothesis: If codon pairs can be selected due to tRNA structural constrains on the ribosome P and A sites then it might be necessary to correct for these codon preferences when studying synonymous mutations.
DNA mutations that do not change the coding amino-acid are many times referred to as "silent mutations", or synonymous mutations, because it is less likely that they will result in a change in function. Synonymous mutations are often considered to be evolutionary neutral and the ratio of non-synonymous substitutions (Ka) to synonymous substitutions (Ks) is used to study sequence evolution. It can be used for example to search for DNA regions targeted by selection (see review and a practical application).
In the last issue of Science Kimchi-Sarfaty and colleagues found a synonymous mutation in a transport protein that has an effect on the protein function. They have shown, at least in cell-lines, that the mutation does not affect mRNA levels nor the produced protein sequence. Finally the authors showed that the mutation might change the protein's conformation by comparing the sensibility of wild type and mutated sequence to trypsin digestion.
The authors speculate that the usage of that particular codon, even if not affecting the coding region, might change the translation rate and folding of the protein. It had already been shown in E. coli that synonymous mutations can affect the in vivo folding of a protein. Here the authors have shown a case where a silent mutation can change the substrate specificity of a transporter.
Because of these codon preferences it is important to adjust for codon selection pressures when studying synonymous substitutions. The codon preferences are usually considered to be due to differences in the pool of the cognate tRNA but other studies have shown that codon bias might arise also by codon context. In E. coli, codon pair preferences, were observed to affect their in vivo translation. Also, these codon pair preferences are species specific and are, at least in part, influenced by nucleotide positions within A-site tRNA sequences.
Hypothesis: If codon pairs can be selected due to tRNA structural constrains on the ribosome P and A sites then it might be necessary to correct for these codon preferences when studying synonymous mutations.
Tuesday, January 23, 2007
System Biology quick links
(via Pierre) BMC System Biology has published their first papers. More or less at the same time the new Systems and Synthetic Biology (published by Springer Netherlands) has started publishing papers. These two journals join IEE Systems Biology and Molecular Systems Biology (Nature/EMBO) as forums to publish works on Systems and Synthetic Biology. All journals (with the exception of IEE Systems Biology) publish in open access or at least (in the case of Systems and Synthetic Biology) offer an open access option.
Some of the talks from the BioSysBio conference are online in Goggle Video.
Here is a nice talk from Alfonso Valencia talking about species co-evolution and a very promising improvement to a sequence based method to predict protein-protein interactions:
(via Pierre) BMC System Biology has published their first papers. More or less at the same time the new Systems and Synthetic Biology (published by Springer Netherlands) has started publishing papers. These two journals join IEE Systems Biology and Molecular Systems Biology (Nature/EMBO) as forums to publish works on Systems and Synthetic Biology. All journals (with the exception of IEE Systems Biology) publish in open access or at least (in the case of Systems and Synthetic Biology) offer an open access option.
Some of the talks from the BioSysBio conference are online in Goggle Video.
Here is a nice talk from Alfonso Valencia talking about species co-evolution and a very promising improvement to a sequence based method to predict protein-protein interactions:
Monday, January 22, 2007
Social gene annotation in Connotea
There has been a lot of excitement over the recent web technological developments. Time magazine has recognized this by announcing that instead of profiling an individual in their annual issue of Person of Year they decided to select You as the most influential group of last year. This "you" refers to everyone that is out there on the web building, interacting, blogging, uploading their videos and pictures for the world to see. As with almost every rising meme, the backlash is inevitable. Some see this web euphoria as little more than global narcissism.
This social web holds some powerful promises of more efficient collaboration but clear examples might still be lacking. Scientists, given our need to communicate and collaborate, are a group of individuals that could do more to take advantage of these tools. Unfortunately we seem to be too unaware and too slow to pick them up.
I have shown before that the accumulating body of knowledge in Connotea, in the form of simple tagging of science papers, can in principle be used to highlight papers of higher impact.
I tough that it could also be possible to mine Connotea to retrieve gene annotations. I tested if manuscripts tagged as "cell-cycle" and "yeast" would contain, in their abstracts, mostly genes names related to cell cycle in yeast. There are currently 38 papers in Connotea tagged as cell-cycle and yeast with an associated Pubmed ID. I used a dictionary of S. cerevisiae gene names obtained from SGD and retrieved the abstracts for the 38 manuscripts using eUtils.
Within these abstracts there were 38 gene names associated by a simple pattern match. To evaluate the performance of this social gene annotation I took from the SGD's slim GO mapping the function and processes associated to these genes. I also included the gene description from gene name registry.
Table 1 - Known GO process/function annotations and gene function description associated to the genes predicted to participate in cell-cycle in yeast by social annotations.
From the 38 genes, 14 (~37%) are annotated in the slim GO annotation as participating in cell-cycle,meiosis or cytokinesis. From the remaining, 15 (39%) have a described function associated to the cell-cycle (ex. G1 cyclin involved in cell cycle progression, expression restricted to mother cells in late G1 as controlled by Swi4p-Swi6p, Swi5p and Ash1p,etc). In total roughly 76% of the gene names obtained are associated to cell-cycle in S. cerevisiae.

This simple test highlights the potential usefulness of social bookmarking of science papers. However it was limited to a very specific field and to a very small number of annotated manuscripts. Hopefully someone can come up with a better way of testing this :).
There has been a lot of excitement over the recent web technological developments. Time magazine has recognized this by announcing that instead of profiling an individual in their annual issue of Person of Year they decided to select You as the most influential group of last year. This "you" refers to everyone that is out there on the web building, interacting, blogging, uploading their videos and pictures for the world to see. As with almost every rising meme, the backlash is inevitable. Some see this web euphoria as little more than global narcissism.
This social web holds some powerful promises of more efficient collaboration but clear examples might still be lacking. Scientists, given our need to communicate and collaborate, are a group of individuals that could do more to take advantage of these tools. Unfortunately we seem to be too unaware and too slow to pick them up.
I have shown before that the accumulating body of knowledge in Connotea, in the form of simple tagging of science papers, can in principle be used to highlight papers of higher impact.
I tough that it could also be possible to mine Connotea to retrieve gene annotations. I tested if manuscripts tagged as "cell-cycle" and "yeast" would contain, in their abstracts, mostly genes names related to cell cycle in yeast. There are currently 38 papers in Connotea tagged as cell-cycle and yeast with an associated Pubmed ID. I used a dictionary of S. cerevisiae gene names obtained from SGD and retrieved the abstracts for the 38 manuscripts using eUtils.
Within these abstracts there were 38 gene names associated by a simple pattern match. To evaluate the performance of this social gene annotation I took from the SGD's slim GO mapping the function and processes associated to these genes. I also included the gene description from gene name registry.
Table 1 - Known GO process/function annotations and gene function description associated to the genes predicted to participate in cell-cycle in yeast by social annotations.
From the 38 genes, 14 (~37%) are annotated in the slim GO annotation as participating in cell-cycle,meiosis or cytokinesis. From the remaining, 15 (39%) have a described function associated to the cell-cycle (ex. G1 cyclin involved in cell cycle progression, expression restricted to mother cells in late G1 as controlled by Swi4p-Swi6p, Swi5p and Ash1p,etc). In total roughly 76% of the gene names obtained are associated to cell-cycle in S. cerevisiae.

This simple test highlights the potential usefulness of social bookmarking of science papers. However it was limited to a very specific field and to a very small number of annotated manuscripts. Hopefully someone can come up with a better way of testing this :).
Thursday, January 18, 2007
Petition for guaranteed public access
(via PLoS publishing blog):
"A group of European organisations - JISC (Joint Information Systems Committee, UK), SURF (Netherlands), SPARC Europe, DFG (Deutsches Forschungsgemeinschaft, Germany), DEFF (Denmark's Electronic Research Library) - have posted a petition to encourage the EC to formally endorse the open access recommendations."
This petition recommends that "any potential 'embargo' on free access should be set at no more than six months following publication" for any EC funded research.
Have a look and sign the petition if you are for it.
(via PLoS publishing blog):
"A group of European organisations - JISC (Joint Information Systems Committee, UK), SURF (Netherlands), SPARC Europe, DFG (Deutsches Forschungsgemeinschaft, Germany), DEFF (Denmark's Electronic Research Library) - have posted a petition to encourage the EC to formally endorse the open access recommendations."
This petition recommends that "any potential 'embargo' on free access should be set at no more than six months following publication" for any EC funded research.
Have a look and sign the petition if you are for it.
Sunday, January 14, 2007
Bio::Blogs# 7 and some quick links
The bioinformatics blog journal Bio::Blogs will have it's 7th edition on the 1st of February. We skipped the January edition because of the holidays. It will be hosted by Paras Chopra on BioHacking blog. Anyone can submit the link to their posts on paras1987 {at} gmail or bioblogs {at} gmail until the end of this month.
Some quick links:
Paras released the source code of a Python program for protein structure prediction.
(via Gerstein' blog) Yale university has a podcast. I wish I could convince EMBL's press office to start blogging and/or a podcast.
(via Deepak) The Science Commons blog announced that the three journals published by EMBO and NPG (EMBO reports, EMBO journal and Molecular Systems Biolgoy) will soon start publishing with a creative commons license. More information on the subject can be found in the EMBO site. In the case of Molecular Systems Biology all articles are published in open access but for EMBO Journal and EMBO reports it looks like the author will decide if they wish to pay an extra fee (2000 euros) to publish in open access. Only the articles published in open access will be published with the creative commons license. Adopting the creative commons license will make re-using their papers much easier, hopefully increasing the usefulness of their content.
(disclaimer: I am currently working for Molecular Systems Biology. All opinions expressed in this blog are my own)
Speaking of re-using content. Alf has set up a mirror site for PLoS One. He called it PLoS Too :) and he is using it to try out some ideas on layout, microformats and features like rating. This is one funny thing about the creative commons license. As long as you give credit to the source you are free to re-use the content. Nothing stops a group of people from setting up a new journal, based on those that are published in creative commons, with a different editorial line. For this particular license you can even try to make some money from re-using the content :).
The bioinformatics blog journal Bio::Blogs will have it's 7th edition on the 1st of February. We skipped the January edition because of the holidays. It will be hosted by Paras Chopra on BioHacking blog. Anyone can submit the link to their posts on paras1987 {at} gmail or bioblogs {at} gmail until the end of this month.
Some quick links:
Paras released the source code of a Python program for protein structure prediction.
(via Gerstein' blog) Yale university has a podcast. I wish I could convince EMBL's press office to start blogging and/or a podcast.
(via Deepak) The Science Commons blog announced that the three journals published by EMBO and NPG (EMBO reports, EMBO journal and Molecular Systems Biolgoy) will soon start publishing with a creative commons license. More information on the subject can be found in the EMBO site. In the case of Molecular Systems Biology all articles are published in open access but for EMBO Journal and EMBO reports it looks like the author will decide if they wish to pay an extra fee (2000 euros) to publish in open access. Only the articles published in open access will be published with the creative commons license. Adopting the creative commons license will make re-using their papers much easier, hopefully increasing the usefulness of their content.
(disclaimer: I am currently working for Molecular Systems Biology. All opinions expressed in this blog are my own)
Speaking of re-using content. Alf has set up a mirror site for PLoS One. He called it PLoS Too :) and he is using it to try out some ideas on layout, microformats and features like rating. This is one funny thing about the creative commons license. As long as you give credit to the source you are free to re-use the content. Nothing stops a group of people from setting up a new journal, based on those that are published in creative commons, with a different editorial line. For this particular license you can even try to make some money from re-using the content :).
Thursday, January 11, 2007
Scientific Journals blog
Blogs have been around for some time. From the wikepedia:
Blogs in science, on the other hand, have only recently become popular. The first two traditional science news journals to pick up the raising interest in scientific blogging were The-Scientist in August 2005, and then Nature in December 2005. Since then many more scientist have picked up blogging for a variety of purposes (see review by Coturnix). The science journals have been slowly reacting. Here is a current list of blogs from science journal blogs or publishing groups. If anyone knows more please leave a comment and I will add them to the list.
Blogs have been around for some time. From the wikepedia:
The term "weblog" was coined by Jorn Barger on 17 December 1997. The short form, "blog," was coined by Peter Merholz, who jokingly broke the word weblog into the phrase we blog in the sidebar of his blog Peterme.com in April or May of 1999.
Blogs in science, on the other hand, have only recently become popular. The first two traditional science news journals to pick up the raising interest in scientific blogging were The-Scientist in August 2005, and then Nature in December 2005. Since then many more scientist have picked up blogging for a variety of purposes (see review by Coturnix). The science journals have been slowly reacting. Here is a current list of blogs from science journal blogs or publishing groups. If anyone knows more please leave a comment and I will add them to the list.
| Journal | Blog |
| The Scientist | |
| Scientific American | |
| Nature publishing group | |
| Nature Journals | |
| Nature Genetics | |
| Nature Neuroscience | |
| Nature Methods | |
| Nature Medicine | |
| Nature News | |
| Chemistry at Nature (portal not journal) | |
| Nature publishing group | |
| Nature publishing group | |
| Nature publishing group | |
| Heredity | |
| Public Library of Science | |
| PLoS Blogs | |
| PLoS publishing | |
| PLoS Technology | |
| PLoS Medicine | |
| The Lancet | |
| Science | The Weblog of Science Magazine"s (stopped) |
Wednesday, January 10, 2007
Science Blogging Anthology 2006
I mentioned before that Coturnix was getting ready a list of blog posts that would go into a science blogging anthology of 2006. Twelve judges have selected 50 blog posts that will be put together in a book. The book will then be published by lulu. The judges were nice enough to select one of my posts to go in the book :)
Opening up the scientific process
I mentioned before that Coturnix was getting ready a list of blog posts that would go into a science blogging anthology of 2006. Twelve judges have selected 50 blog posts that will be put together in a book. The book will then be published by lulu. The judges were nice enough to select one of my posts to go in the book :)
Opening up the scientific process
Saturday, January 06, 2007
Science Blogging Anthology
Coturnix from a Blog Around the Clock is organizing a science blogging anthology. I missed it during the Christmas holidays but the results are due in a couple of days. Here is the list of posts that got nominated and are now being evaluated. One of my posts made the nomination list :) cool.
Last month Roland Krause said that this type of vanity posts (like blog carnivals) are similar to spam blogs. I actually think that there is value in carnivals and other equivalent content promotion activities. They create a cheap reward system that motivates people to produce more and better content. They also provide with a layer of quality rating even if, in the case of carnivals, the posts that are submitted are self contributed. Bloggers tend to submit their best content to the carnivals.
On a related note but with a very different opinion, here is a rant on Web 2.0 And Narcissism (via Rough Type):
Coturnix from a Blog Around the Clock is organizing a science blogging anthology. I missed it during the Christmas holidays but the results are due in a couple of days. Here is the list of posts that got nominated and are now being evaluated. One of my posts made the nomination list :) cool.
Last month Roland Krause said that this type of vanity posts (like blog carnivals) are similar to spam blogs. I actually think that there is value in carnivals and other equivalent content promotion activities. They create a cheap reward system that motivates people to produce more and better content. They also provide with a layer of quality rating even if, in the case of carnivals, the posts that are submitted are self contributed. Bloggers tend to submit their best content to the carnivals.
On a related note but with a very different opinion, here is a rant on Web 2.0 And Narcissism (via Rough Type):
"What he's getting at is that this whole Web 2.0, social networking, virtual community business is essentially a pornography of the self—a projected, fictionalized self that is then worshipped by the slightly less-perfect self."
Thursday, January 04, 2007
Specificity and Evolvability in Protein Interaction Networks
I finally have the opportunity to blog about some of what I have worked on during last year. It has been published in PLoS Computational Biology and is freely available here in the early online release format (still in the original ugly format :). One way to use our blogs may be to add some depth to the papers that we published. Something like the extras we get when we buy the DVD of a movie ;)
Main conclusions
- Protein interactions can change at a fast rate of 1E-5 interactions per protein pair per million years
- Binding specificity is a strong determining of binding specificity with more promiscuous binding proteins having a higher rate of change of interactions.
- Human proteins involved in immune response, transport and establishment of localization, show signs of positive selection for change of interactions.
The making of
We had been using comparative genomics to search for conserved putative protein binding sites. These very conserved putative target sites were very likely to be experimentally known target sites but many other known binding sites seamed not to be so conserved. This was what got us started thinking about the evolution of these protein interactions and what might determine the rate at which interactions are gained and lost during evolution. The analysis was mostly inspired on the nice work of Andreas Wagner that first proposed a rate for the addition of new interactions in S. cerevisiae. We have tried to build on this by analyzing different species and determining also what protein properties might determine the rate at which interactions are gained and lost in evolution.
More than nodes and edges
One of the main conclusions from this work was that binding specificity also determines the rate of change of interactions during evolution. More promiscuous binding proteins not only have many binding partners but they also tend to change partners faster during evolution. To establish a proxy for binding specificity we have used structural information from the iPFAM database. In essence we have considered that protein domains that have been seen in contact with many different other domains would be more promiscuous. In general we observed that proteins containing these promiscuous domains had a high rate of change of interactions (see figure below).
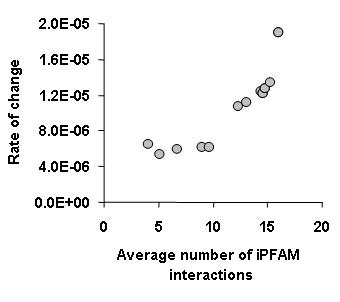
This highlights something that I had stressed before, that it is important to consider protein interaction networks as more than nodes and edges. Another recent paper has also shown that it is possible and useful to use the accumulating structural information available in the PDB to obtain a more accurate representation of protein interaction networks. Philip M. Kim and co workers from the Gerstein lab (blog, webpage) published a study in Science were they have used also the iPFAM database to curate all the S. cerevisie interactions and to discriminate between interactions that use the same or different binding interfaces. With this information the authors distinguish between hubs that tend to interact with their partners mostly trough one interface or trough many interfaces. They have shown that the multi interface hubs are more restricted in evolution and more likely to be essential than single interface hubs. They have a website with presentations and additional data for this paper.
In the pipeline
To be submitted soon (hopefully), are some collaborations on how to use structural information to predict protein-protein binding specificity. With these collaborations I finish my thesis (still waiting for the defense). During the next couple of months I am off to search for a lab to work as a postdoc.
I finally have the opportunity to blog about some of what I have worked on during last year. It has been published in PLoS Computational Biology and is freely available here in the early online release format (still in the original ugly format :). One way to use our blogs may be to add some depth to the papers that we published. Something like the extras we get when we buy the DVD of a movie ;)
Main conclusions
- Protein interactions can change at a fast rate of 1E-5 interactions per protein pair per million years
- Binding specificity is a strong determining of binding specificity with more promiscuous binding proteins having a higher rate of change of interactions.
- Human proteins involved in immune response, transport and establishment of localization, show signs of positive selection for change of interactions.
The making of
We had been using comparative genomics to search for conserved putative protein binding sites. These very conserved putative target sites were very likely to be experimentally known target sites but many other known binding sites seamed not to be so conserved. This was what got us started thinking about the evolution of these protein interactions and what might determine the rate at which interactions are gained and lost during evolution. The analysis was mostly inspired on the nice work of Andreas Wagner that first proposed a rate for the addition of new interactions in S. cerevisiae. We have tried to build on this by analyzing different species and determining also what protein properties might determine the rate at which interactions are gained and lost in evolution.
More than nodes and edges
One of the main conclusions from this work was that binding specificity also determines the rate of change of interactions during evolution. More promiscuous binding proteins not only have many binding partners but they also tend to change partners faster during evolution. To establish a proxy for binding specificity we have used structural information from the iPFAM database. In essence we have considered that protein domains that have been seen in contact with many different other domains would be more promiscuous. In general we observed that proteins containing these promiscuous domains had a high rate of change of interactions (see figure below).

This highlights something that I had stressed before, that it is important to consider protein interaction networks as more than nodes and edges. Another recent paper has also shown that it is possible and useful to use the accumulating structural information available in the PDB to obtain a more accurate representation of protein interaction networks. Philip M. Kim and co workers from the Gerstein lab (blog, webpage) published a study in Science were they have used also the iPFAM database to curate all the S. cerevisie interactions and to discriminate between interactions that use the same or different binding interfaces. With this information the authors distinguish between hubs that tend to interact with their partners mostly trough one interface or trough many interfaces. They have shown that the multi interface hubs are more restricted in evolution and more likely to be essential than single interface hubs. They have a website with presentations and additional data for this paper.
In the pipeline
To be submitted soon (hopefully), are some collaborations on how to use structural information to predict protein-protein binding specificity. With these collaborations I finish my thesis (still waiting for the defense). During the next couple of months I am off to search for a lab to work as a postdoc.
Monday, December 25, 2006
Publish your GreaseMonkey scripts
I knew it was possible to use GreaseMonkey scripts to change webpages to better suit our needs. I tried it once to get blog comments about scientific papers to show up in journal websites. Pierre and Stew have been creating several interesting scripts for postgenomic and connotea. What I did not know was that one could actually publish these scripts. I am all for publishing of smaller and finner grained scientific content but I have to say that this paper seemed like little more than a big blog post. Should we try to publish this type of work ?
Anyway :) ho ho ho , merry xmas everyone
I knew it was possible to use GreaseMonkey scripts to change webpages to better suit our needs. I tried it once to get blog comments about scientific papers to show up in journal websites. Pierre and Stew have been creating several interesting scripts for postgenomic and connotea. What I did not know was that one could actually publish these scripts. I am all for publishing of smaller and finner grained scientific content but I have to say that this paper seemed like little more than a big blog post. Should we try to publish this type of work ?
Anyway :) ho ho ho , merry xmas everyone
Friday, December 22, 2006
PLoS One is up and running
I will add this blog's "voice" to the many that have already announced the start of PLoS One. This journal is a very much needed experiment in science communication online. It is being built from scratch to take advantage of the internet as the medium unlike many other journals. As Deepak mentioned in his blog, the success of One will depend mostly on us, the users, in our interest to participate with our own insight. The PLoS One team will have to worry about creating interesting reward systems around the journal to help boost participation.
I have only played around on the site for a short while but there are a couple of features that I hope that will implement in the short term.
- One that I was hoping to find at start was some form of gateways or portals for areas. The only subject navigation available seams to be the links on the right side.
- I also did not find any kind of rating system. To boost participation I think people have to start trying out simple participation systems and rating is the easiest one.
- I guess it would also be nice to have some kind of track back system or some other way to let comments come in from blog post. This would be nice for bloggers but i have to admit that few people would care :) .
There were two papers (from the same group) that caught my attention but I did not have time to read them:
Control of Canalization and Evolvability by Hsp90
Modularity and Intrinsic Evolvability of Hsp90-Buffered Change
I will add this blog's "voice" to the many that have already announced the start of PLoS One. This journal is a very much needed experiment in science communication online. It is being built from scratch to take advantage of the internet as the medium unlike many other journals. As Deepak mentioned in his blog, the success of One will depend mostly on us, the users, in our interest to participate with our own insight. The PLoS One team will have to worry about creating interesting reward systems around the journal to help boost participation.
I have only played around on the site for a short while but there are a couple of features that I hope that will implement in the short term.
- One that I was hoping to find at start was some form of gateways or portals for areas. The only subject navigation available seams to be the links on the right side.
- I also did not find any kind of rating system. To boost participation I think people have to start trying out simple participation systems and rating is the easiest one.
- I guess it would also be nice to have some kind of track back system or some other way to let comments come in from blog post. This would be nice for bloggers but i have to admit that few people would care :) .
There were two papers (from the same group) that caught my attention but I did not have time to read them:
Control of Canalization and Evolvability by Hsp90
Modularity and Intrinsic Evolvability of Hsp90-Buffered Change
Tags: plos,plosone,open science
Tuesday, December 05, 2006
Second (hellish) Life
I had a walk around Second Nature inside Second Life. I had tried SL before some months ago and this time the experience was much worse. My avatar keeps freezing when I touch objects or when I stand around idle for some time. Some other times the avatar just freezes in flight and continues in the same direction until I quit. It was just unusable.
Here is a picture from an 3D cell in the Second Nature island.

A feeble attempt to build something for the EMBL online symposium

I had a walk around Second Nature inside Second Life. I had tried SL before some months ago and this time the experience was much worse. My avatar keeps freezing when I touch objects or when I stand around idle for some time. Some other times the avatar just freezes in flight and continues in the same direction until I quit. It was just unusable.
Here is a picture from an 3D cell in the Second Nature island.

A feeble attempt to build something for the EMBL online symposium

Tags: second life,science
Monday, December 04, 2006
Modularity and Evolvability
The First Online EMBL PhD Symposium as started today with several media files available for viewing and commenting. There is an IRC channel open for discussion.
Participants can comment or add their own content to the site. I have put up a small presentation relating modularity and evolvability in proteins, language, software and the scientific process. I am no expert in any of these fields so view these analogies with a very critical eye :).
Full screen view can be access from the slideshare site
The First Online EMBL PhD Symposium as started today with several media files available for viewing and commenting. There is an IRC channel open for discussion.
Participants can comment or add their own content to the site. I have put up a small presentation relating modularity and evolvability in proteins, language, software and the scientific process. I am no expert in any of these fields so view these analogies with a very critical eye :).
Full screen view can be access from the slideshare site
Saturday, December 02, 2006

The 6th edition of Bio::Blogs is up in Nodalpoint. Many thanks to Greg for setting it up. This edition marks half a year of Bio::Blogs and it is dedicated to conference blogging, probably one of the best examples of the usefulness of science blogging.
The 7th edition will probably be scheduled to February to skip the holiday season and Paras Chopra has volunteered to host it.
Tags: bio::blogs,blog canivals,bioblogs
Monday, November 27, 2006
EMBL online PhD symposium

(via Notes from the biomass)
EMBL is organizing the 1st PhD online symposium (4-8 December). It will be fully online, available for free to anyone. Participants can register to participate in the discussions and watch the presentations when they are made available. The users can create pages to share posters, talks or anything that might be relevant to the conference. In all, it could be a good playground to try out new ideas for online conferences. We could ask Nature to broadcast it in Nature island inside second life ? :)
Register, create your pages, share some thoughts. The topics are:
Career Development
Omics Session / Systems Biology
Scientific Communication 2.0
Konrad is one of the organizers.

(via Notes from the biomass)
EMBL is organizing the 1st PhD online symposium (4-8 December). It will be fully online, available for free to anyone. Participants can register to participate in the discussions and watch the presentations when they are made available. The users can create pages to share posters, talks or anything that might be relevant to the conference. In all, it could be a good playground to try out new ideas for online conferences. We could ask Nature to broadcast it in Nature island inside second life ? :)
Register, create your pages, share some thoughts. The topics are:
Career Development
Omics Session / Systems Biology
Scientific Communication 2.0
Konrad is one of the organizers.
Tags: open science, EMBL,conference
Tuesday, November 21, 2006
Connotea tag:evolution citation report
What is a scientific journal ? One possible definition could be - a content provider that filters and selects scientific content appropriate (of interest) to a particular group of people. Currently, journals select papers based on the decisions of a small group of people, maybe one or two editors supported by a few referees. The internet allows for alternative methods to select and filter content based potentially on the knowledge of a larger group of people. Eventually, these methods might one day replace the expansive editorial procedures now in place in most journals, but before that happens these approaches have to be evaluated. Also, even if we don't use these methods to replace current editorial procedures, they can be used to help us highlight the most interesting works published in certain fields.
So, why not consider tags in social bookmarking services like Connotea as scientific journals ? Here is the journal Connotea tag:evolution. I took from Connotea yesterday (21/11/2006) all papers tagged with the tag "evolution" , that were published in 2003 or 2004 (85 papers). I used the web of science to get the number of citations of each of these papers (see figure below). This was unfortunately done one by one. I am thinking of scripting some tool to do it automatically but if someone knows a better way please let me know.
 As expected the most represented journals are some of the journals with higher visibility but still more than 50% of the manuscripts were tagged from more specialized journals.
As expected the most represented journals are some of the journals with higher visibility but still more than 50% of the manuscripts were tagged from more specialized journals.
 So how does the average number of citations per paper of this "new journal" compare with well established journals ?
So how does the average number of citations per paper of this "new journal" compare with well established journals ?
 Although Connotea Evolution is low volume compared to other journals it does have a higher average citation per paper than journals such as Nature and Science. I did not separate potential non citable items from any of the groups so it should be a reasonable fair comparison.
Although Connotea Evolution is low volume compared to other journals it does have a higher average citation per paper than journals such as Nature and Science. I did not separate potential non citable items from any of the groups so it should be a reasonable fair comparison.
I think this suggests that we should evaluate potential mechanism to guide us to interesting scientific content. By itself, these evaluations might establish a form of reward for the community to come up with more sophisticated tools. It is important therefore to carefully pick the measurements. I used the citations per paper but others might be more adequate.
Any group of people can use the internet to re-group scientific content (specially if it is open access) into "journals" of potentially more value than those currently available.
What is a scientific journal ? One possible definition could be - a content provider that filters and selects scientific content appropriate (of interest) to a particular group of people. Currently, journals select papers based on the decisions of a small group of people, maybe one or two editors supported by a few referees. The internet allows for alternative methods to select and filter content based potentially on the knowledge of a larger group of people. Eventually, these methods might one day replace the expansive editorial procedures now in place in most journals, but before that happens these approaches have to be evaluated. Also, even if we don't use these methods to replace current editorial procedures, they can be used to help us highlight the most interesting works published in certain fields.
So, why not consider tags in social bookmarking services like Connotea as scientific journals ? Here is the journal Connotea tag:evolution. I took from Connotea yesterday (21/11/2006) all papers tagged with the tag "evolution" , that were published in 2003 or 2004 (85 papers). I used the web of science to get the number of citations of each of these papers (see figure below). This was unfortunately done one by one. I am thinking of scripting some tool to do it automatically but if someone knows a better way please let me know.
 As expected the most represented journals are some of the journals with higher visibility but still more than 50% of the manuscripts were tagged from more specialized journals.
As expected the most represented journals are some of the journals with higher visibility but still more than 50% of the manuscripts were tagged from more specialized journals. So how does the average number of citations per paper of this "new journal" compare with well established journals ?
So how does the average number of citations per paper of this "new journal" compare with well established journals ? Although Connotea Evolution is low volume compared to other journals it does have a higher average citation per paper than journals such as Nature and Science. I did not separate potential non citable items from any of the groups so it should be a reasonable fair comparison.
Although Connotea Evolution is low volume compared to other journals it does have a higher average citation per paper than journals such as Nature and Science. I did not separate potential non citable items from any of the groups so it should be a reasonable fair comparison.I think this suggests that we should evaluate potential mechanism to guide us to interesting scientific content. By itself, these evaluations might establish a form of reward for the community to come up with more sophisticated tools. It is important therefore to carefully pick the measurements. I used the citations per paper but others might be more adequate.
Any group of people can use the internet to re-group scientific content (specially if it is open access) into "journals" of potentially more value than those currently available.
Monday, November 20, 2006
Journals Proliferate
I only noticed today BMC also had a Systems Biology journal. There are no papers yet but the editorial board looks interesting enough. From the types of articles they expect I think they will take a lot of bioinformatics related manuscripts. This adds to two other Systems Biology journals that I am aware of: Molecular Systems Biology (also open access) and IET Systems Biology.
(via BioHacking) On a more creative note, here is the Journal of Visualized Experiments. It is a "journal" of recorded experiments that should help others learn protocols with the aid of videos. Currently the submissions are subjected only to editorial evaluation and are expected to get published in about 14 days. They plan to apply for listing in PubMed and other databases.
The videos are accompanied by a very short written explanation and are tagged for searching. There are no comments or RSS features that I could see.
Nature Methods and Nature Protocols should give this a try. By the way, Nature Protocols also publishes protocols in bioinformatics, and Nature Methods started a blog (Methagora)
With this continuing expansion of journals in all publishing houses aren't we quickly reaching a point when manuscripts will be the scarce resource ? I really hope someone develops nicer tools to suggest communications to read based on my interests. Is there a place for researchers whose job is just to associate and shuttle communications around ? Tagging communications as FOR_SMITH_J to show up in some reader with a comment: "solves your problem X".
I only noticed today BMC also had a Systems Biology journal. There are no papers yet but the editorial board looks interesting enough. From the types of articles they expect I think they will take a lot of bioinformatics related manuscripts. This adds to two other Systems Biology journals that I am aware of: Molecular Systems Biology (also open access) and IET Systems Biology.
(via BioHacking) On a more creative note, here is the Journal of Visualized Experiments. It is a "journal" of recorded experiments that should help others learn protocols with the aid of videos. Currently the submissions are subjected only to editorial evaluation and are expected to get published in about 14 days. They plan to apply for listing in PubMed and other databases.
The videos are accompanied by a very short written explanation and are tagged for searching. There are no comments or RSS features that I could see.
Nature Methods and Nature Protocols should give this a try. By the way, Nature Protocols also publishes protocols in bioinformatics, and Nature Methods started a blog (Methagora)
With this continuing expansion of journals in all publishing houses aren't we quickly reaching a point when manuscripts will be the scarce resource ? I really hope someone develops nicer tools to suggest communications to read based on my interests. Is there a place for researchers whose job is just to associate and shuttle communications around ? Tagging communications as FOR_SMITH_J to show up in some reader with a comment: "solves your problem X".
Chris Surridge explains PLoS ONE
As part of the OpenWetWare's Seminar Series on Open Science, here is Chris Surridge explaining PLoS ONE. Some random things I remember:
- No-one reads full table of contents anymore so why not create a journal with broad scope ? (I do read full table of contents of many journals, it's the first thing I do in the morning)
- They are aiming at a very high volume (hundreds of manuscripts)
- The journal will probably have portals for subject areas
- Anyone is free to reuse the open access content, so anyone could in theory be an editor by reusing open access content and focusing it for a particular target audience.
- They might also pool in papers from other open access journals
- There will be probably a karma system to rate the contributions
- The system of having different versions of the same manuscript will not be on the first version of the journal.
- The journal might be up on the 29th of this month (not sure yet)
- Chris Surridge moves around too much :)
Tags: plos,plosone,open science
Tuesday, November 14, 2006
iTOL - Interactive Tree Of Life
The Bork group published recently a revised tree of life in Science. You can have a better look at the tree using this nice interactive tree viewer that they put up on the web (publication).

You can zoom in and out, re-root, swap branches, and export the trees. You can also upload your own trees and annotations to have them drawn by the viewer. Hovering over the species brings up a box with some information. Did you ever wonder who donated DNA for the human sequencing project ? The Bork group has found out for us:

The Bork group published recently a revised tree of life in Science. You can have a better look at the tree using this nice interactive tree viewer that they put up on the web (publication).

You can zoom in and out, re-root, swap branches, and export the trees. You can also upload your own trees and annotations to have them drawn by the viewer. Hovering over the species brings up a box with some information. Did you ever wonder who donated DNA for the human sequencing project ? The Bork group has found out for us:

Monday, November 13, 2006
PLoS redesigns, PLoS ONE soon
Go have a look at the PLoS websites, they have been redesigned. I like the look but the only notable changes are a box "From the Blogosphere" that currently links to the Open Access News blog (I wonder why:) and a link to "Readers Respond" on the left, that should put more emphasis on user participation. The PLoS Medicine site has some notable differences. First they include the PLoS Medicine blog on the home page and the "From the Blogosphere" links to a Guardian article. I guess that these are customizable and left to the editors to use to point out interesting things related to the journal or field. Now ... when will PLoS Comp Bio start a blog ?
Still no journal is taking in blog comments. It would be easy to use Postgenomic's index or trackbacks to let readers comments papers trough their blogs.
There is also an editorial in PLoS Biology about PLoS ONE: "ONE for All: The Next Step for PLoS"
The only new thing I got from the editorial was the concept of portals withing PLoS ONE. I would say it sounds a bit like the Nature Gateways, an area for the aggregation of papers and other resources related to a particular field or project. Sounds like a good idea. Again, they mention that at the start it will look like any other journal and that they will build on it in time, so I don't expect much in the launch day.
Go have a look at the PLoS websites, they have been redesigned. I like the look but the only notable changes are a box "From the Blogosphere" that currently links to the Open Access News blog (I wonder why:) and a link to "Readers Respond" on the left, that should put more emphasis on user participation. The PLoS Medicine site has some notable differences. First they include the PLoS Medicine blog on the home page and the "From the Blogosphere" links to a Guardian article. I guess that these are customizable and left to the editors to use to point out interesting things related to the journal or field. Now ... when will PLoS Comp Bio start a blog ?
Still no journal is taking in blog comments. It would be easy to use Postgenomic's index or trackbacks to let readers comments papers trough their blogs.
There is also an editorial in PLoS Biology about PLoS ONE: "ONE for All: The Next Step for PLoS"
The only new thing I got from the editorial was the concept of portals withing PLoS ONE. I would say it sounds a bit like the Nature Gateways, an area for the aggregation of papers and other resources related to a particular field or project. Sounds like a good idea. Again, they mention that at the start it will look like any other journal and that they will build on it in time, so I don't expect much in the launch day.
Tags: plos,plos one,open access
Bio::Blogs#6 @ Nodalpoint
Greg has volunteered to host the 6th edition of Bio:Blogs at Nodalpoint (half a year already). It should take place on the 1st of December. Anyone that wants to participate just has to point out an interesting bioinformatics related blog post by email to the usual bioblogs _at_ gmail _dot_ com.
Some links:
(via postgenomic and Clinical Cases and Images) The Lancet started a blog (creatively called The Lancet blog) . If it funny that some of the oldest journals are testing new ideas in publishing leaving some of the more recent ones lagging behind.
Here is another bioinformatics related blog. From a research associate at Newcastle University, interested in neuroscience.
Greg has volunteered to host the 6th edition of Bio:Blogs at Nodalpoint (half a year already). It should take place on the 1st of December. Anyone that wants to participate just has to point out an interesting bioinformatics related blog post by email to the usual bioblogs _at_ gmail _dot_ com.
Some links:
(via postgenomic and Clinical Cases and Images) The Lancet started a blog (creatively called The Lancet blog) . If it funny that some of the oldest journals are testing new ideas in publishing leaving some of the more recent ones lagging behind.
Here is another bioinformatics related blog. From a research associate at Newcastle University, interested in neuroscience.
Tags: science,bioblogs,thelancet,scienceblogs
Friday, November 10, 2006
MeeboMe to
Fallowing the tip from Deepak I decided to give MeeboMe a try here in the blog.
Fell free to interrupt my thesis writing :) by writing something there on the box.
Fallowing the tip from Deepak I decided to give MeeboMe a try here in the blog.
Fell free to interrupt my thesis writing :) by writing something there on the box.
Tags: meebome,meebo,blog things
Thursday, November 09, 2006
Personal fabrication - Update
Several years ago I read a book by Nicholas Negroponte entitled "Being Digital". It was a really great introduction to the digital revolution and provided with a glimpse to some of the changes that are still ongoing in our society. I find fascinating the ability that some technologies have to change so much our ways of living, opening up with a simple stroke so many new possibilities. The digital revolution was one of these events. An apparently simple concept of having information coded in a digital format that can be transported anywhere at near instantaneous speeds. Now we have the internet that as provided with so many wonderful advantages. I can learn anything I want, provided I have time. I can collaborate with people I have never even met before to build new things. It is difficult even for me to imagine living in an unwired world (but that might be a bit geeky thing to say :).
I read today this profile on Neil Gershenfeld and his work on home manufacturing at MIT and I though that this would also be a very empowering technology. He is pushing the concept of personal fabrication, enabling anyone to easily create any physical thing. He wants to bring the same speed to making atoms that we have today for bits. You could grab a funny chair design that you made in your computer, send it to your friends by email and they would open it .. very literally by "printing" it and sitting on it to try it out.
I guess there are lots of details to think about, but generally the idea sounds fun.
I was thinking that some companies should have a go at this already. I am not really into design but I guess that several companies today focus on constantly renewing their stock with new designs and trend setting ideas. Why not have an Ikea section of "do your own" furniture. Something like social web manufacturing of clothes, accessories, furniture, etc.
Also, why not have collection of the most used objects in Second Life, built every quarter or half a year. Second Life authors would have to agree and probably negotiate a cut of the profit.
Here is someone building a guitar :)
Where is the print button to get one of those right now?
Update:
I am really behind on this meme. I few more clicks and I found this blog post in MIT Advertising lab. From there I found this company that specializes in bringing virtual objects to life.
A blog post by Ian Hughes on home fabrication.
A wiki page fab@home with instructions to make "fabbers", machines that can make almost anything, right on your desktop.
Several years ago I read a book by Nicholas Negroponte entitled "Being Digital". It was a really great introduction to the digital revolution and provided with a glimpse to some of the changes that are still ongoing in our society. I find fascinating the ability that some technologies have to change so much our ways of living, opening up with a simple stroke so many new possibilities. The digital revolution was one of these events. An apparently simple concept of having information coded in a digital format that can be transported anywhere at near instantaneous speeds. Now we have the internet that as provided with so many wonderful advantages. I can learn anything I want, provided I have time. I can collaborate with people I have never even met before to build new things. It is difficult even for me to imagine living in an unwired world (but that might be a bit geeky thing to say :).
I read today this profile on Neil Gershenfeld and his work on home manufacturing at MIT and I though that this would also be a very empowering technology. He is pushing the concept of personal fabrication, enabling anyone to easily create any physical thing. He wants to bring the same speed to making atoms that we have today for bits. You could grab a funny chair design that you made in your computer, send it to your friends by email and they would open it .. very literally by "printing" it and sitting on it to try it out.
I guess there are lots of details to think about, but generally the idea sounds fun.
I was thinking that some companies should have a go at this already. I am not really into design but I guess that several companies today focus on constantly renewing their stock with new designs and trend setting ideas. Why not have an Ikea section of "do your own" furniture. Something like social web manufacturing of clothes, accessories, furniture, etc.
Also, why not have collection of the most used objects in Second Life, built every quarter or half a year. Second Life authors would have to agree and probably negotiate a cut of the profit.
Here is someone building a guitar :)
Where is the print button to get one of those right now?
Update:
I am really behind on this meme. I few more clicks and I found this blog post in MIT Advertising lab. From there I found this company that specializes in bringing virtual objects to life.
A blog post by Ian Hughes on home fabrication.
A wiki page fab@home with instructions to make "fabbers", machines that can make almost anything, right on your desktop.
Tags: Personal fabrication
Sunday, November 05, 2006
Activating RNAs (RNAa) - another twist
A paper published online in PNAS described a possible new form of regulation of gene expression by small RNAs. The authors found that small dsRNAs can also serve as transcriptional activators. They showed that the mechanism for gene expression activation also prefers dsRNAs of ~21 nt in size and requires the Argonaute 2 protein.
The story was covered by a news article in Science. From the news article it looks like there is some fear that the result might be due to indirect effects (inactivation of other genes leading to gene expression activation). Also, some wonder why this as not been picked up before by other studies.
I am very tempted right now to get some of the high-throughput screens of RNAi to try dig out some more examples that might have gone unnoticed (if I actually had any time to do it :).
A paper published online in PNAS described a possible new form of regulation of gene expression by small RNAs. The authors found that small dsRNAs can also serve as transcriptional activators. They showed that the mechanism for gene expression activation also prefers dsRNAs of ~21 nt in size and requires the Argonaute 2 protein.
The story was covered by a news article in Science. From the news article it looks like there is some fear that the result might be due to indirect effects (inactivation of other genes leading to gene expression activation). Also, some wonder why this as not been picked up before by other studies.
I am very tempted right now to get some of the high-throughput screens of RNAi to try dig out some more examples that might have gone unnoticed (if I actually had any time to do it :).
Wednesday, November 01, 2006
Bio::Blogs#5
The November edition of our bioinformatics blog journal is up at Chris' blog. This edition is mostly focused on potencially interesting tools to use, including Zotero, a bibliography manager Firefox extension and Bioclipse, an open-source workbench for bio/chemi-informatics.
I forgot to ask Chris to link to the bio::blogs icon challenge. I guess we can run it for another month. If you don't like these two icons and you think you can do better give it a try :) until next month.
We are in need of a volunteer to host the December edition. If you run a bioinformatics related blog and are interested send in a link to your blog to bioblogs at gmail.com or leave a comment below.
The November edition of our bioinformatics blog journal is up at Chris' blog. This edition is mostly focused on potencially interesting tools to use, including Zotero, a bibliography manager Firefox extension and Bioclipse, an open-source workbench for bio/chemi-informatics.
I forgot to ask Chris to link to the bio::blogs icon challenge. I guess we can run it for another month. If you don't like these two icons and you think you can do better give it a try :) until next month.
We are in need of a volunteer to host the December edition. If you run a bioinformatics related blog and are interested send in a link to your blog to bioblogs at gmail.com or leave a comment below.
Subscribe to:
Comments (Atom)