Social network dynamics in a conference setting
(disclaimer: This was not peer reviewed and is not serious at all :)
To study the dynamics in social network topology we decided evaluate how some nodes (also called humans) interact in defined experimental conditions. We used the scientific meeting setting that we think can serve as a model for this type of studies. We observed human-human interactions during the meeting breaks by taking snapshots and calculating inter-human distances. We defined an arbitrary cut-off to determine the binary interactions between all the humans present in the study.
The first analysis we preformed was under the so call "conference breaks" model where our nodes are allowed to interact for brief time intervals after being subjected lengthy lectures.
We observed an interesting clustered network topology that can be described with a power law distribution. Most nodes in the network have few interactions while a small fractions of humans was found to consistently interact with a large number of other nodes. We found also some nodes that did not show any interactions in our studies even when several "conference breaks" were preformed. We believe that these could be pseudo-humans that were included in our study by mistake. These pseudo-humans might be on the way to extinction from the humeone.
Having built this network of human-human interaction on a large scale we decided to investigate what human properties might be correlated with human hubs. We used previous large-scale studies of human properties like height, gender and number of papers published to test this.
We show here that although gender shows a significant correlation with human hubness, the best predictor for hubs in the conference breaks networks is actually number of papers published. We tried to refine this further by introducing a new human measurement we call "hypeness". Hypeness of a human was calculated as a modification of the number of papers published weighted by the impact factor of the journals where the papers were published and also the number of times cited in popular media articles. We show here that hypeness does significant better at predicting hub nodes in this network.
Given that networks are dynamic we set out to map the changes in network structure with time. To simulate this we perturbed the gathering using a small-compound (EtOH) that we administered in liquid form. With time we observed a noticeable change in the network. Although the overall topological properties were maintained, the nature of the hubs changed dramatically. In this new network state that we call the "drunk" state, the best predictor for the highly connected hubs is clearly gender. We believe this clearly proves that social networks in conference settings are very dynamic with time.
To prove that gender was indeed the best indicator of hubness and not some strange artifact we used deletions studies. Random female nodes where struck with a sudden case of "sleepiness" and the perturbed network was observed. We show here that random female deletion leads to a rapid collapse of the network. The same is not observed with random deletion of the hypest nodes, proving our initial proposition.
Tuesday, September 05, 2006
Propagation of Errors in Review Articles
Thomas J. Katz signs a small letter in Science warning us about the propagation of errors in review articles. The author gives a scary example of an incorrect citation propagated through 9 reviews (if I counted correctly). The cited paper does not contain the experiment that all the reviews mention and as it seems it was actually never published anywhere. Very scary.
As more science moves online with more individual voices, will this propagation of errors be accentuated or reduced?
Thomas J. Katz signs a small letter in Science warning us about the propagation of errors in review articles. The author gives a scary example of an incorrect citation propagated through 9 reviews (if I counted correctly). The cited paper does not contain the experiment that all the reviews mention and as it seems it was actually never published anywhere. Very scary.
As more science moves online with more individual voices, will this propagation of errors be accentuated or reduced?
Tags: reviews, science, error propagation
How to recognize you have become senior faculty
I am back from holidays and trying to plow trough the RSS feeds/content alerts that accumulated in these two weeks. I might post on couple of things that catch my eye.
Here is a funny editorial from Gregory A Petsko talking about the project to sequence Homo neanderthalensis.
The editorial is actually more about senior faculty members and in particular how to identify one:
- You are senior faculty if you can actually remember when more than 10% of submitted grants got funded.
- You are senior faculty if you can remember when there was only one Nature.
You are senior faculty if you still get a lot of invitations to meetings, but they're all to deliver after-dinner talks.
- You are senior faculty if students sometimes ask you if you ever heard Franklin in person, and they mean Benjamin, not Aretha.
- You are senior faculty if a junior colleague wants to know what it was like before computers, and you can tell her.
- You are senior faculty when the second joint on the little finger of your left hand is the only joint that isn't stiff at the end of a long seminar.
- You are senior faculty if you sleep through most of those long seminars.
- You are senior faculty if you visit the Museum of Natural History, and the dummies in the exhibit of Stone Age man all remind you of people you went to school with.
- You are senior faculty if you find yourself saying "Back in my day" or "When I was your age" at least twice a week.
- You are senior faculty if you actually know what investigator-initiated, hypothesis-driven research means.
- You are senior faculty if you occasionally think that maybe you should attend a faculty meeting once in a while.
- You are senior faculty when your CV includes papers you can't remember writing.
I am back from holidays and trying to plow trough the RSS feeds/content alerts that accumulated in these two weeks. I might post on couple of things that catch my eye.
Here is a funny editorial from Gregory A Petsko talking about the project to sequence Homo neanderthalensis.
The editorial is actually more about senior faculty members and in particular how to identify one:
- You are senior faculty if you can actually remember when more than 10% of submitted grants got funded.
- You are senior faculty if you can remember when there was only one Nature.
You are senior faculty if you still get a lot of invitations to meetings, but they're all to deliver after-dinner talks.
- You are senior faculty if students sometimes ask you if you ever heard Franklin in person, and they mean Benjamin, not Aretha.
- You are senior faculty if a junior colleague wants to know what it was like before computers, and you can tell her.
- You are senior faculty when the second joint on the little finger of your left hand is the only joint that isn't stiff at the end of a long seminar.
- You are senior faculty if you sleep through most of those long seminars.
- You are senior faculty if you visit the Museum of Natural History, and the dummies in the exhibit of Stone Age man all remind you of people you went to school with.
- You are senior faculty if you find yourself saying "Back in my day" or "When I was your age" at least twice a week.
- You are senior faculty if you actually know what investigator-initiated, hypothesis-driven research means.
- You are senior faculty if you occasionally think that maybe you should attend a faculty meeting once in a while.
- You are senior faculty when your CV includes papers you can't remember writing.
Monday, September 04, 2006
Bio::Blogs #3
The third edition of Bio::Blogs was released a couple of days ago in business|bytes|genes|molecules.
I particularly enjoyed the nice discussions going on in evolgen , about the rifts in scientific communities and in Neil's blog regarding structural genomics data.
The next Bio::Blogs will be edited by Sandra Porter. Send your links and offers to host future editions to bioblogs{at}gmail.com.
The third edition of Bio::Blogs was released a couple of days ago in business|bytes|genes|molecules.
I particularly enjoyed the nice discussions going on in evolgen , about the rifts in scientific communities and in Neil's blog regarding structural genomics data.
The next Bio::Blogs will be edited by Sandra Porter. Send your links and offers to host future editions to bioblogs{at}gmail.com.
Tags: bio::blogs, carnivals, blog carnivals
Sunday, August 27, 2006
Bio::Blogs #3 - call for submissions
The third edition of Bio::Blogs will be up on the 1st of September, edited by mndoci. Send your submissions to the usual email bioblogs {at} gmail.com. There were few submissions so far. It might be a slow month if a lot of people took some time off, or maybe most people are waiting for the last day as usual.
I am back from holidays, trying to digest the emails and RSS feeds accumulated in two weeks. I miss the beach already :).
The third edition of Bio::Blogs will be up on the 1st of September, edited by mndoci. Send your submissions to the usual email bioblogs {at} gmail.com. There were few submissions so far. It might be a slow month if a lot of people took some time off, or maybe most people are waiting for the last day as usual.
I am back from holidays, trying to digest the emails and RSS feeds accumulated in two weeks. I miss the beach already :).
Tags: blog carnivals, bio::blogs
Tuesday, August 15, 2006
Interactome Networks conference
I am going for a two week holidays tomorrow. I really need some boring relaxing days by the beach without thinking to much about anything :).
After that I am going to a conference, Interactome Networks on the Wellcome Trust Genome Campus in Hinxton, UK. I will give a short talk about my last project - "Specificity and evolvability in eukaryotic protein interaction networks". I will try to blog some of the talks, either here or in Nodalpoint.
Here is the current list of talks and posters. If by any chance you are not going and are particularly interested in some of the titles let me know and I will try to have a look.
I am going for a two week holidays tomorrow. I really need some boring relaxing days by the beach without thinking to much about anything :).
After that I am going to a conference, Interactome Networks on the Wellcome Trust Genome Campus in Hinxton, UK. I will give a short talk about my last project - "Specificity and evolvability in eukaryotic protein interaction networks". I will try to blog some of the talks, either here or in Nodalpoint.
Here is the current list of talks and posters. If by any chance you are not going and are particularly interested in some of the titles let me know and I will try to have a look.
Tags: networks, conference blogging
Sunday, August 13, 2006
Science Foo Camp
I am to sleepy to post a coherent account of what happened today at scifoo, I'll try to do it tomorrow in Nodalpoint. It's an amazing mixture of people that they gathered here, social/bio/physics/computer scientists, science fiction writers (futurists?), journal editors, open access and open science advocates and googlers.
To ilustrate how chaotic the meeting has been, here is a picture of the session planing board they put up.

So anyone can just grab a pen and fill up a slot. The only problem so far was actually having too many parallel interesting sessions.
I am to sleepy to post a coherent account of what happened today at scifoo, I'll try to do it tomorrow in Nodalpoint. It's an amazing mixture of people that they gathered here, social/bio/physics/computer scientists, science fiction writers (futurists?), journal editors, open access and open science advocates and googlers.
To ilustrate how chaotic the meeting has been, here is a picture of the session planing board they put up.

So anyone can just grab a pen and fill up a slot. The only problem so far was actually having too many parallel interesting sessions.
Tags: scifoo, conference blogging
Thursday, August 10, 2006
Science Foo Camp
I am going to California this weekend to attend the Science Foo Camp. An event organized by Nature/O'Reilly and hosted by Google. Yeap I get to visit the Googleplex :) (I admit it am I geek). It has been fun to see the event getting set up. It started with a wiki page seeded with some ideas from O'Reilly and some instructions. Then everyone started editing their bios and suggesting/offering talks.
I will be blogging my impressions of the event over at Nodalpoint but probably a good way to keep track of the event is to take a look at the scifoo tag in connotea.
(official announcement)
I am going to California this weekend to attend the Science Foo Camp. An event organized by Nature/O'Reilly and hosted by Google. Yeap I get to visit the Googleplex :) (I admit it am I geek). It has been fun to see the event getting set up. It started with a wiki page seeded with some ideas from O'Reilly and some instructions. Then everyone started editing their bios and suggesting/offering talks.
I will be blogging my impressions of the event over at Nodalpoint but probably a good way to keep track of the event is to take a look at the scifoo tag in connotea.
(official announcement)
Tags: scifoo, science, conference
Wednesday, August 09, 2006
Blogging science .. something like science
(via Open Reading frame and Uncertain Principles) This is what happens when you spend to much time in the lab. Dylan Stiles reports on his analysis of ... his own ear wax?! Creative post to say the least. :).
(via Open Reading frame and Uncertain Principles) This is what happens when you spend to much time in the lab. Dylan Stiles reports on his analysis of ... his own ear wax?! Creative post to say the least. :).
Tags: fun in lab, NMR
Tuesday, August 08, 2006
Identifying protein-protein interfaces
There were several interesting papers on protein interaction in the last week. For example, in PLoS Computational Biology, Kim et al described an improvement to a method that classifies interaction interfaces according to their geometry. The results of the analysis are available on their site SCOPPI.
It seems reasonable to believe that there is a limited number of protein interaction types (predicted to be around 10 thousand), much the same way that there is probably a limited number of folds used in nature. This database, along side others like the iPFAM database, provide templates on which to model other protein interactions with reasonable homology. As first proposed by Aloy and Russell, if we know that two proteins interact, for example with a yeast-two-hybrid experiment we might be able to use these databases to identity and model the interaction interface between the two proteins.
Here is an example of two complexes taken from the paper. The nodes and edges view hides the fact that RBP and RCC1 interact through different interfaces.
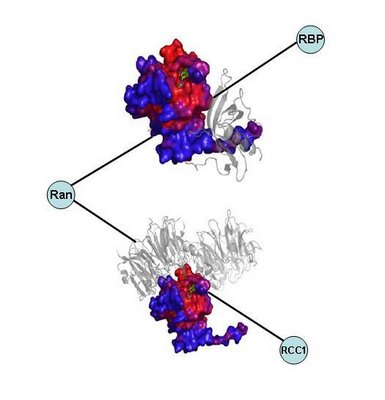
Although is has been very useful to look at protein networks as of bunch of nodes connected by edges for some analysis it would be much more informative to know what are the interacting interfaces.
The authors used the database to claim that:
- hub proteins interact with proteins using many distinct faces (what they actually show is that domains that interact with many different other domain types have more distint faces);
- two thirds of gene fusions conserve the binding orientation;
- the apparent poor conservation of interfaces is due to the diversity of interactions and partners (in my opinion it is more a suggestion that proof);
- the interfaces common to archae, bacteria and eukaryotes and mostly symmetric homo-dimers, suggesting that asymmetric and hetero interactions evolved from symmetric homo-dimers.
<speculation> Maybe if we could convert the current human interactome into more than nodes and edges we could try to see if some of disease causing polymorphisms can be explained by how they affect the interfaces. Maybe we could use this to do interaction KOs instead of whole proteins </speculation>
There were several interesting papers on protein interaction in the last week. For example, in PLoS Computational Biology, Kim et al described an improvement to a method that classifies interaction interfaces according to their geometry. The results of the analysis are available on their site SCOPPI.
It seems reasonable to believe that there is a limited number of protein interaction types (predicted to be around 10 thousand), much the same way that there is probably a limited number of folds used in nature. This database, along side others like the iPFAM database, provide templates on which to model other protein interactions with reasonable homology. As first proposed by Aloy and Russell, if we know that two proteins interact, for example with a yeast-two-hybrid experiment we might be able to use these databases to identity and model the interaction interface between the two proteins.
Here is an example of two complexes taken from the paper. The nodes and edges view hides the fact that RBP and RCC1 interact through different interfaces.

Although is has been very useful to look at protein networks as of bunch of nodes connected by edges for some analysis it would be much more informative to know what are the interacting interfaces.
The authors used the database to claim that:
- hub proteins interact with proteins using many distinct faces (what they actually show is that domains that interact with many different other domain types have more distint faces);
- two thirds of gene fusions conserve the binding orientation;
- the apparent poor conservation of interfaces is due to the diversity of interactions and partners (in my opinion it is more a suggestion that proof);
- the interfaces common to archae, bacteria and eukaryotes and mostly symmetric homo-dimers, suggesting that asymmetric and hetero interactions evolved from symmetric homo-dimers.
<speculation> Maybe if we could convert the current human interactome into more than nodes and edges we could try to see if some of disease causing polymorphisms can be explained by how they affect the interfaces. Maybe we could use this to do interaction KOs instead of whole proteins </speculation>
Saturday, August 05, 2006
PLoS ONE is accepting manuscripts - Update
(via PLoS blog and Open) The new PLoS ONE journal is now accepting manuscripts for review.They are still taking care of a few bugs and at this moments I could only use the site with Internet Explorer.
We can know have a look at the editorial board and the journal policies. There are some hints of how the user comments and ratings are going to work.
Most journals have some form of funneling, either by perceived impact or subject scope. It is going to be interesting to see what happens with PLoS ONE, given that there is no editorial selection on these criteria.
Update - It is actually working fine with Firefox1.5. When I was seeing it before the the page layout was misbehaving. Sorry for the confusion.
(via PLoS blog and Open) The new PLoS ONE journal is now accepting manuscripts for review.
We can know have a look at the editorial board and the journal policies. There are some hints of how the user comments and ratings are going to work.
Most journals have some form of funneling, either by perceived impact or subject scope. It is going to be interesting to see what happens with PLoS ONE, given that there is no editorial selection on these criteria.
Update - It is actually working fine with Firefox1.5. When I was seeing it before the the page layout was misbehaving. Sorry for the confusion.
Tags: open access, PLoS, PLoS ONE
Friday, August 04, 2006
Meta blogging
Neil Saunders' blog as moved to a new home. Fabrice Jossinet is back blogging about RNA and bioinformatics in Propeller Twist.
If you are interested in RNA and microbiology go check out the new blog of Rosie Redfield. She runs a a microbiology lab at the University of British Columbia and will be blogging about their current research.
If you are interested in a better way to track the comments and responses to your comments in other blogs have a look at coComments. You can have an RSS feed and/or a box in your blog with your comments and responses to the comments that you wish to track.
Neil Saunders' blog as moved to a new home. Fabrice Jossinet is back blogging about RNA and bioinformatics in Propeller Twist.
If you are interested in RNA and microbiology go check out the new blog of Rosie Redfield. She runs a a microbiology lab at the University of British Columbia and will be blogging about their current research.
If you are interested in a better way to track the comments and responses to your comments in other blogs have a look at coComments. You can have an RSS feed and/or a box in your blog with your comments and responses to the comments that you wish to track.
Tags: "meta blogging"
Wednesday, August 02, 2006
Tangled Bank #59
The latest issue of Tangled Bank is up at Science and Reason. It is the first time I participate with a submission. If you are into physics the author of the blog, Charles Daney, is considering starting a physics carnival.
The latest issue of Tangled Bank is up at Science and Reason. It is the first time I participate with a submission. If you are into physics the author of the blog, Charles Daney, is considering starting a physics carnival.
Tags: blog carnivals, science
Tuesday, August 01, 2006
Bio::Blogs#2
The second edition of Bio::Blogs is up at Neil's blog. Go check it out and participate with your insightful comments :). There is a lot of conference blogging on this issue, a nice way to get updates on the conferences you might have missed.
I am happy to say that we have volunteers for the next two editions. Deepak offered to host for September 1st and Sandra Porter will host the October 1st edition.
The second edition of Bio::Blogs is up at Neil's blog. Go check it out and participate with your insightful comments :). There is a lot of conference blogging on this issue, a nice way to get updates on the conferences you might have missed.
I am happy to say that we have volunteers for the next two editions. Deepak offered to host for September 1st and Sandra Porter will host the October 1st edition.
Tags: blog carnivals, bio::blogs
Saturday, July 29, 2006
The likelihood that two proteins interact might depend on the proteins' age - part 2
Abstract
It has been previously shown [1] that S. cerevisiae proteins preferentially interact with proteins of the same estimated likely time of origin. Using a similar approach but focusing on a less broad evolutionary time span I observed that the likelihood for protein interactions depends on the proteins’ age. I had show this previously for the interactome of S. cerevisiae [2] and here I extend the analysis to show that the same is also observed for the interactome of H. sapiens. Importantly the observation does not depend on the experimental method used since removing the yeast-two-hybrid interactions does not alter the result.
Methods and Results
Protein-protein interactions for H.sapiens were obtained from the Human Protein Reference database and from two high-throughput studies excluding any interactions derived from protein complexes. I considered only proteins that were represented in this interactome (i.e. with one or more interactions).
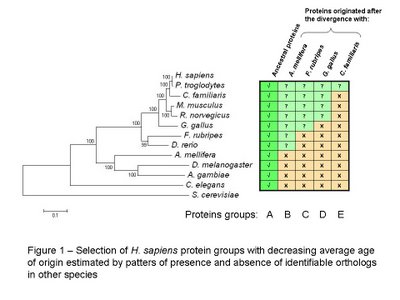
As before I created groups of H. sapiens proteins with different average age using the reciprocal best blast hit method to determine the most likely ortholog in eleven other eukaryotic species (see figure 1 for species names). For a more detailed description of the group selection and the construction of the phylogenetic tree please see the previous post [2].
It is important to note that the placement of C. familiaris does not correspond with other published phylogenetic trees it might be due to the proteins selected for the tree construction. I should consider using different combinations of ancestral proteins to check the robustness of the tree.
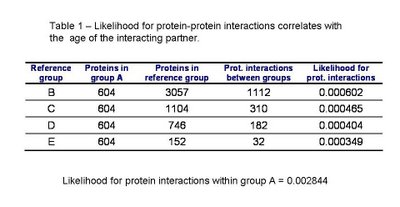
In table 1 we can see the likelihood for protein interactions to occur within the ancestral proteins of group A and between the ancestral proteins and other groups of decreasing average age. As published by Qin et al. and as I had observed before for S. cerevisiae, the interactions within groups of the same age (group A) are more likely than between groups of proteins of different times of origin. Also, the likelihood for a protein to interact with an ancestral protein depends on the age of this protein. Confirming the pervious observation that the younger the protein is the less likely it is to interact with an ancestral protein.
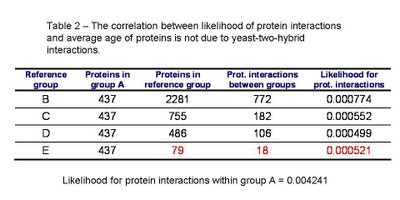
I redid the analysis excluding yeast-two-hybrid interactions from the dataset. As it can be see in table 2, the results are qualitatively the same. There is a small increase in the likelihood of interaction with the ancestral proteins for the youngest group (highlighted in red in table 2) that is likely due to lack of data.
Caveats and possible continuations
I still have to test the statistical significance of these observations and control for possible other effects like protein size and protein expression that could explain these results.
I am interested in continuing this further as an open project. Fallowing the suggestion of Roland Krause I will soon start a wiki page to dump the data bits accumulated for open discussion. Hopefully more people will join in and maybe we can together shape up a small communication.
[1]Qin H, Lu HH, Wu WB, Li WH. Evolution of the yeast protein interaction network. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12820-4. Epub 2003 Oct 13
[2]Beltrao. P The likelihood that two proteins interact might depend on the proteins' age Blog post
Abstract
It has been previously shown [1] that S. cerevisiae proteins preferentially interact with proteins of the same estimated likely time of origin. Using a similar approach but focusing on a less broad evolutionary time span I observed that the likelihood for protein interactions depends on the proteins’ age. I had show this previously for the interactome of S. cerevisiae [2] and here I extend the analysis to show that the same is also observed for the interactome of H. sapiens. Importantly the observation does not depend on the experimental method used since removing the yeast-two-hybrid interactions does not alter the result.
Methods and Results
Protein-protein interactions for H.sapiens were obtained from the Human Protein Reference database and from two high-throughput studies excluding any interactions derived from protein complexes. I considered only proteins that were represented in this interactome (i.e. with one or more interactions).
As before I created groups of H. sapiens proteins with different average age using the reciprocal best blast hit method to determine the most likely ortholog in eleven other eukaryotic species (see figure 1 for species names). For a more detailed description of the group selection and the construction of the phylogenetic tree please see the previous post [2].
It is important to note that the placement of C. familiaris does not correspond with other published phylogenetic trees it might be due to the proteins selected for the tree construction. I should consider using different combinations of ancestral proteins to check the robustness of the tree.

In table 1 we can see the likelihood for protein interactions to occur within the ancestral proteins of group A and between the ancestral proteins and other groups of decreasing average age. As published by Qin et al. and as I had observed before for S. cerevisiae, the interactions within groups of the same age (group A) are more likely than between groups of proteins of different times of origin. Also, the likelihood for a protein to interact with an ancestral protein depends on the age of this protein. Confirming the pervious observation that the younger the protein is the less likely it is to interact with an ancestral protein.

I redid the analysis excluding yeast-two-hybrid interactions from the dataset. As it can be see in table 2, the results are qualitatively the same. There is a small increase in the likelihood of interaction with the ancestral proteins for the youngest group (highlighted in red in table 2) that is likely due to lack of data.

Caveats and possible continuations
I still have to test the statistical significance of these observations and control for possible other effects like protein size and protein expression that could explain these results.
I am interested in continuing this further as an open project. Fallowing the suggestion of Roland Krause I will soon start a wiki page to dump the data bits accumulated for open discussion. Hopefully more people will join in and maybe we can together shape up a small communication.
[1]Qin H, Lu HH, Wu WB, Li WH. Evolution of the yeast protein interaction network. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12820-4. Epub 2003 Oct 13
[2]Beltrao. P The likelihood that two proteins interact might depend on the proteins' age Blog post
Tags: protein interactions, evolution, networks
Friday, July 28, 2006
Binding specificity and complexity
There is a paper out in PNAS about the distribution of free energy of binding for the yeast-two-hybrid datasets. Although I still have to dig into the model they used I found the result quite interesting. They observe that the average binding energy decreases with cellular complexity.
They have some sentences in there that made my hairs stand like: "more evolved organisms have weaker binary protein-protein binding". What does "more evolved" mean ? Also on figure 4 of the paper they plot miu (a parameter related to the average binding energy) over divergence times without saying what species they are comparing.
This result fits well with another paper published a while ago in PLoS Comp Bio about protein family expansions and complexity. Christine Vogel and Cyrus Chothia show (among other things) what protein domains expansion best correlate with complexity. They used cell numbers as a proxy for species complexity. If you look at the top of the list (in table 2) you can find several of the peptide binding domains, know to be of low specificity, given that they do not require a folded structure to interact with.
What I would like to know is the correlation between binding affinity and binding specificity. For example SH2 domains bind much more tightly than SH3 domains although they are both not very specific binding domains. Maybe in general it could be said that average lower binding affinities correspond to lower average binding specificity.
Why would complexity correlate with binding specificity ? I think one important factor is cellular size. An increase is size has allowed for exploration of spacial factors in determining cellular response. Specificity of binding in the real cell (not in binary assays) is determined also by localization at sub cellular structures.
One practical reminder coming from this is that even if we have the perfect method to determine biophysical binding specificity we are still going to get poor results if we cannot predict all other components that will determine if the two proteins will bind or not (i.e localization, expression).
There is a paper out in PNAS about the distribution of free energy of binding for the yeast-two-hybrid datasets. Although I still have to dig into the model they used I found the result quite interesting. They observe that the average binding energy decreases with cellular complexity.
They have some sentences in there that made my hairs stand like: "more evolved organisms have weaker binary protein-protein binding". What does "more evolved" mean ? Also on figure 4 of the paper they plot miu (a parameter related to the average binding energy) over divergence times without saying what species they are comparing.
This result fits well with another paper published a while ago in PLoS Comp Bio about protein family expansions and complexity. Christine Vogel and Cyrus Chothia show (among other things) what protein domains expansion best correlate with complexity. They used cell numbers as a proxy for species complexity. If you look at the top of the list (in table 2) you can find several of the peptide binding domains, know to be of low specificity, given that they do not require a folded structure to interact with.
What I would like to know is the correlation between binding affinity and binding specificity. For example SH2 domains bind much more tightly than SH3 domains although they are both not very specific binding domains. Maybe in general it could be said that average lower binding affinities correspond to lower average binding specificity.
Why would complexity correlate with binding specificity ? I think one important factor is cellular size. An increase is size has allowed for exploration of spacial factors in determining cellular response. Specificity of binding in the real cell (not in binary assays) is determined also by localization at sub cellular structures.
One practical reminder coming from this is that even if we have the perfect method to determine biophysical binding specificity we are still going to get poor results if we cannot predict all other components that will determine if the two proteins will bind or not (i.e localization, expression).
TOPAZ and PLoS ONE
According to the PLoS blog the new PLoS ONE will be accepting submissions soon. I guess they will at the same time release the TOPAZ system that will likely be available here.
According to the PLoS blog the new PLoS ONE will be accepting submissions soon. I guess they will at the same time release the TOPAZ system that will likely be available here.
"TOPAZ will serve the rapidly growing demand for sophisticated tools and resources to read and use the scientific and medical literature, allowing scholarly publishers, societies, universities, and research communities to publish open access journals economically and efficiently."
Tags: open access, plos, topaz
Sunday, July 23, 2006
Opening up the scientific process
During my stay at the EMBL, for the past couple of years, it already happened more than once that people I know have been scooped. This simple means that all the hard work that they have been doing was already done by someone else that manage to publish it a bit sooner and therefore limited severely the usefulness of their discoveries. Very few journals are interested in publishing research that merely confirms other published results.
From talking to other people, I have come to accept that scooping is a part of science. There is no other possible conclusion from this but to accept that the scientific process is very flawed. We should not be wasting resources literally racing with each other to be the first person to discover something. When you try to explain to non-scientist, that it is very common to have 3 or 4 labs doing exactly the same thing they usually have a hard time integrating this with their perception of science as the pursue of knowledge trough collaboration.
I am probably naïve given that I am only doing this for a couple of years but I don’t pretend to say that we do not need competition in science. We need to keep each other in check exactly because lack of competition leads to waste of resources. I would argue however that right now the scientific process is creating competition at wrong levels decreasing the potential productivity.
So how do we work and what do we aim to produce? We are in the business of producing manuscripts accepted in peer reviewed journals. To have competition there most be a scarce element. In our case the limited element is the attention of fellow scientist. Given that scientist’s attention is scarce we all compete for the limited number of time that researchers have to read papers every week. So the good news is that the system tends to give credit to high quality manuscripts. This means that research projects and ongoing results should be absolutely confidential and everything should be focused in getting that Science or Nature paper.
I found a beautiful drawing of an iceberg (used here with permission from the author, David Fierstein) that I think illustrates the problem we have today by focusing the competition on the manuscripts. Only a small fraction of the research process is in view.

Wouldn’t it be great if we could find a way to make most of the scientific process public but at the same time guaranty some level of competition? What I think we could do would be to define steps in the process that we could say are independent, which can work as modules. Here I mean module in the sense of a black box with inputs and outputs that we wire together without caring too much on how the internals of the boxes work. I am thinking these days about these modules and here is a first draft of what this could look like:
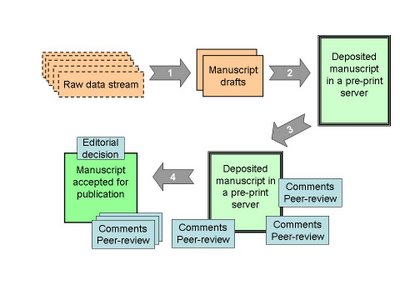
The data streams would be, as the name suggests, a public view of the data being produced by a group or individual researcher. Blogs are a simple way this could be achieved today (see for example this blog). The manuscripts could be built in wikis by selection of relevant data bits from the streams that fit together to answer an interesting question. This is where I propose that the competition would come in. Only those relevant bits of data that better answer the question would be used. The authors of the manuscript would be all those that contributed data bits or in some other way contributed for the manuscript creation. In this way all the data would be public and still a healthy level of competition would be maintained.
The rest of the process could go on in public view. Versions of the manuscript deemed stable could be deposited in a pre-print server and comments and peer review would commence. Latter there could still be another step of competition to get the paper formally accepted in a journal.
One advantage of this is that it is not a revolution of the scientific process. People could still work in their normal research environment closed within their research groups. This is just a model of how we could extend the system to make it mostly open and public. The technologies are all here: structured blogging for the data streams, wikis for the manuscripts and online communities to drive the research agendas.
I think it is important to view the scientific process as a group of modules also because it allows us latter to think of different ways to wire the modules together. Increasing the modularity should permit us to innovate. For example we can latter think of ways that the data streams are brought together to answer questions, etc.
During my stay at the EMBL, for the past couple of years, it already happened more than once that people I know have been scooped. This simple means that all the hard work that they have been doing was already done by someone else that manage to publish it a bit sooner and therefore limited severely the usefulness of their discoveries. Very few journals are interested in publishing research that merely confirms other published results.
From talking to other people, I have come to accept that scooping is a part of science. There is no other possible conclusion from this but to accept that the scientific process is very flawed. We should not be wasting resources literally racing with each other to be the first person to discover something. When you try to explain to non-scientist, that it is very common to have 3 or 4 labs doing exactly the same thing they usually have a hard time integrating this with their perception of science as the pursue of knowledge trough collaboration.
I am probably naïve given that I am only doing this for a couple of years but I don’t pretend to say that we do not need competition in science. We need to keep each other in check exactly because lack of competition leads to waste of resources. I would argue however that right now the scientific process is creating competition at wrong levels decreasing the potential productivity.
So how do we work and what do we aim to produce? We are in the business of producing manuscripts accepted in peer reviewed journals. To have competition there most be a scarce element. In our case the limited element is the attention of fellow scientist. Given that scientist’s attention is scarce we all compete for the limited number of time that researchers have to read papers every week. So the good news is that the system tends to give credit to high quality manuscripts. This means that research projects and ongoing results should be absolutely confidential and everything should be focused in getting that Science or Nature paper.
I found a beautiful drawing of an iceberg (used here with permission from the author, David Fierstein) that I think illustrates the problem we have today by focusing the competition on the manuscripts. Only a small fraction of the research process is in view.

Wouldn’t it be great if we could find a way to make most of the scientific process public but at the same time guaranty some level of competition? What I think we could do would be to define steps in the process that we could say are independent, which can work as modules. Here I mean module in the sense of a black box with inputs and outputs that we wire together without caring too much on how the internals of the boxes work. I am thinking these days about these modules and here is a first draft of what this could look like:

The data streams would be, as the name suggests, a public view of the data being produced by a group or individual researcher. Blogs are a simple way this could be achieved today (see for example this blog). The manuscripts could be built in wikis by selection of relevant data bits from the streams that fit together to answer an interesting question. This is where I propose that the competition would come in. Only those relevant bits of data that better answer the question would be used. The authors of the manuscript would be all those that contributed data bits or in some other way contributed for the manuscript creation. In this way all the data would be public and still a healthy level of competition would be maintained.
The rest of the process could go on in public view. Versions of the manuscript deemed stable could be deposited in a pre-print server and comments and peer review would commence. Latter there could still be another step of competition to get the paper formally accepted in a journal.
One advantage of this is that it is not a revolution of the scientific process. People could still work in their normal research environment closed within their research groups. This is just a model of how we could extend the system to make it mostly open and public. The technologies are all here: structured blogging for the data streams, wikis for the manuscripts and online communities to drive the research agendas.
I think it is important to view the scientific process as a group of modules also because it allows us latter to think of different ways to wire the modules together. Increasing the modularity should permit us to innovate. For example we can latter think of ways that the data streams are brought together to answer questions, etc.
Tags: open science, science2.0, science
Friday, July 21, 2006
Bio::Blogs #2 - call for submissions
(via Nodalpoint) This is just a quick reminder that we have 10 days to submit links to the second edition of Bio::Blogs. You can send your suggestions to bioblogs {at} gmail.com. Also if you wish to host future editions send in a quick email with your name and link to your blog to the same email address.
(via Nodalpoint) This is just a quick reminder that we have 10 days to submit links to the second edition of Bio::Blogs. You can send your suggestions to bioblogs {at} gmail.com. Also if you wish to host future editions send in a quick email with your name and link to your blog to the same email address.
Tags: bio blogs, blog carnival, bioinformatics
Monday, July 17, 2006
Conference on Systems Biology of Mammalian Cells
There was a Systems Biology conference here in Heidelberg last week. For those interested the recorded talks are now available on their site. There is a lot of interesting things about the behavior of network motifs and about network modeling.
There was a Systems Biology conference here in Heidelberg last week. For those interested the recorded talks are now available on their site. There is a lot of interesting things about the behavior of network motifs and about network modeling.
Tags: systems biology, conference, webcast, science
Sunday, July 16, 2006
Blog changes
Notes from the Biomass is back again in a new website. I was cleaning the links on the blog to better reflect what I am actually reading and while I was at it I changed the template. It looks better in IE than in Firefox but I really don't have the time nor the ability to work on a good design.
Notes from the Biomass is back again in a new website. I was cleaning the links on the blog to better reflect what I am actually reading and while I was at it I changed the template. It looks better in IE than in Firefox but I really don't have the time nor the ability to work on a good design.
Tuesday, July 11, 2006
Defrag my life
I am taking the week to visit my former lab in Aveiro, Portugal where I spent one year trying to understand how a codon reassignment occurred in the evolutionary past of C. albicans. This was where I first got into Perl and the wonders of comparative genomics.
It brings back a lot of memories every time I come back to one of the cities I lived in before (6 cities and counting) and I sometimes wonder if it is really necessary for scientists to live such fragmented lives.
reboot, restart, new program.
The regular programming will return soon :).
I am taking the week to visit my former lab in Aveiro, Portugal where I spent one year trying to understand how a codon reassignment occurred in the evolutionary past of C. albicans. This was where I first got into Perl and the wonders of comparative genomics.
It brings back a lot of memories every time I come back to one of the cities I lived in before (6 cities and counting) and I sometimes wonder if it is really necessary for scientists to live such fragmented lives.
reboot, restart, new program.
The regular programming will return soon :).
Tuesday, July 04, 2006
Re: The ninth wave
I usually reed Gregory A Petsko' comments and editorials in Genome Biology that are unfortunately only available with subscription. In the last edition of the journal he wrote a comment entitled "The ninth wave". I have lived most of my life 10min away from the Atlantic ocean and at least to my recollection we used to talk about the 7th wave not the ninth as the biggest wave in a set of waves, but this it not the point :).
Petsko argues that the increase of free access to information on the web and of computer savvy investigators presents a clear danger of a flood of useless correlations hinting at potential discoveries never followed by careful experimental work:
This reminded me of a review I read recently from Andy Clark (via Evolgen). Andy Clark talks about the huge increase of researchers in comparative genomics:
I have a feeling that this is the opinion of a lot of researchers. There is this generalized consensus that people working on computational biology have it easy. Sitting at the computer all day, inventing correlations with other people's data.
Maybe some people feel this way because it is relatively fast to go from idea to result using computers if you have in a mind clearly what you want to test while the experimental work certainly takes longer.
Why should I re-do the experimental work if I can answer a question that I think is interesting using available information ? I should be criticized if I try to overinterpret the results, if the methods used are not appropriate or if the question is not relevant but I should not be criticized for looking for an answer the fastest way I can.
I usually reed Gregory A Petsko' comments and editorials in Genome Biology that are unfortunately only available with subscription. In the last edition of the journal he wrote a comment entitled "The ninth wave". I have lived most of my life 10min away from the Atlantic ocean and at least to my recollection we used to talk about the 7th wave not the ninth as the biggest wave in a set of waves, but this it not the point :).
Petsko argues that the increase of free access to information on the web and of computer savvy investigators presents a clear danger of a flood of useless correlations hinting at potential discoveries never followed by careful experimental work:
Computational analysis of someone else's data, on the other hand, always produces results, and all too often no one but the cognoscenti can tell if these results mean anything.
This reminded me of a review I read recently from Andy Clark (via Evolgen). Andy Clark talks about the huge increase of researchers in comparative genomics:
...one of its worst disasters is that it has created a hoard of genomics investigators who think that evolutionary biology is just fun, speculative story telling. Sadly, much of the scientific publication industry seems to respond to the herd as much as it does to scientific rigor, and so we have a bit of a mess on our hands.
I have a feeling that this is the opinion of a lot of researchers. There is this generalized consensus that people working on computational biology have it easy. Sitting at the computer all day, inventing correlations with other people's data.
Maybe some people feel this way because it is relatively fast to go from idea to result using computers if you have in a mind clearly what you want to test while the experimental work certainly takes longer.
Why should I re-do the experimental work if I can answer a question that I think is interesting using available information ? I should be criticized if I try to overinterpret the results, if the methods used are not appropriate or if the question is not relevant but I should not be criticized for looking for an answer the fastest way I can.
Tags: bioinformatics, meme
Monday, July 03, 2006
Journal policies on preprint servers (2)
Recently I did a survey on the different journal policies regarding preprint servers. I am interested in this because I feel it is important to separate the peer review process from the time-stamping (submission) of a scientific communication. Establishing this separation allows for exploration of alternative and parallel ways of determining the value of a scientific communication. This is only possible if journals accept manuscripts previously deposited in pre-print servers.
Today I received the answer from Bioinformatics:
If you also think that this model, already very established in physics and maths, is useful you can also sent some mails to your journals of interest to enquire about their policies. If enough authors voice their interest there will be more journals accepting manuscripts from pre-print servers.
I think we are now lacking a biomedical preprint server. The Genome Biology journal served until early this year also as a preprint server but they discontinued this practice. Maybe arxiv could expand to include biomedical manuscripts (they already accept quantitative biology manuscripts) .
Recently I did a survey on the different journal policies regarding preprint servers. I am interested in this because I feel it is important to separate the peer review process from the time-stamping (submission) of a scientific communication. Establishing this separation allows for exploration of alternative and parallel ways of determining the value of a scientific communication. This is only possible if journals accept manuscripts previously deposited in pre-print servers.
Today I received the answer from Bioinformatics:
"The Executive Editors have advised that we will allow authors to submit manuscripts to a preprint archive."
If you also think that this model, already very established in physics and maths, is useful you can also sent some mails to your journals of interest to enquire about their policies. If enough authors voice their interest there will be more journals accepting manuscripts from pre-print servers.
I think we are now lacking a biomedical preprint server. The Genome Biology journal served until early this year also as a preprint server but they discontinued this practice. Maybe arxiv could expand to include biomedical manuscripts (they already accept quantitative biology manuscripts) .
Tags: preprint server, bioinformatics, peer review, science
Saturday, July 01, 2006
Bio::Blogs # 1
An editorial of sorts
Welcome to the first edition of Bio::Blogs, a blog carnival covering all subjects related to bioinformatics and computational biology. The main objectives of Bio::Blogs are, in my opinion, to help nit together the bioinformatics blogging community and to showcase some interesting posts on these subjects to other communities. Hopefully it will serve as incentive for other people in the area to start their own blogs and to join in the conversation.
I get to host this edition and I decided to format it more or less like a journal with three sections:1) Conference reports; 2) Primers and reviews; 3) Blog articles. I think this reflects also my opinion on what could be a future role of these carnivals, to serve as a path for certification of scientific content parallel to the current scientific journals.
Given that there were so few submissions I added some links myself. Hopefully in the next editions we can get some more publicity and participation :). Talking about future editions, the second edition of Bio::Blogs will be hosted by Neil and we have now a full month to make something up in our blogs and submit the link to bioblogs{at}gmail{dot}com.
Conference Reports
I selected a blog post from Alf describing what was discussed in a recent conference dedicated to Data Webs. There is a lot of information about potential ways to deal with the increase of data submitted all over the web in many different formats. I remember seeing the advert for this conference and I was intrigued to see Philip Bourne, the editor-in-chief of PLoS Computational Biology, among the speakers. I see know that he is involved in publishing tools under development in PLoS.
Primers & Reviews
Stew from Flags and Lollipops sent in this link to a review on the use of bioinformatics to hunt for disease related genes. He highlights a series of tools and methods that can be used to prioritize candidate genes for experimental validation.
Neil, the next host of Bio::Blogs spent some time with the BioPerl package called Bio::Graphics. He dedicated a blog entry to explain how to create graphics for your results with this package. He gives examples on how to make graphic representations of sequences mapped with blast hits and phosphorylation sites.
Chris, a usual around Nodalpoint, nominated a post in Evolgen:
Evolgen has an interesting post about the relative importance (and interest in) cis and trans acting genetic variation in evo-devo. A lot of (computational) energy has thus far been expended in finding regulatory motifs close to genes (ie, within promoter regions), and conserved elements in non-coding sequences. Rather predictably, cis-acting variants have received the lion's share of attention, probably because they present a more tractable problem. The post deals with work from the evo-devo and comparative genomics fields, but these problems have also been attacked from within-species variation perspectives, particularly the genetics of gene expression. But that's next month's post...
Blog articles
I get to link to my last post. I present some very preliminary results on the influence of protein age on the likelihood of protein-protein interactions. Have fun pointing out all the likely flaws in reasoning and hopefully useful ways to build on it.
To wrap things up here is an announcement by Pierre of a possibly useful applet implementing a Genetic Programming Algorithm. If you ever wanted to play around with genetic programming you can have a go with his applet.
That is it for this month. It is a short Bio::Blogs but I hope you find some of these things useful. Don’t forget to submit the links for the next edition before the end of July. Neil will take up the editorial role for #2 in his blog. If you know of a nice symbol that we might use for Bio::Blogs sent it in as well.
An editorial of sorts
Welcome to the first edition of Bio::Blogs, a blog carnival covering all subjects related to bioinformatics and computational biology. The main objectives of Bio::Blogs are, in my opinion, to help nit together the bioinformatics blogging community and to showcase some interesting posts on these subjects to other communities. Hopefully it will serve as incentive for other people in the area to start their own blogs and to join in the conversation.
I get to host this edition and I decided to format it more or less like a journal with three sections:1) Conference reports; 2) Primers and reviews; 3) Blog articles. I think this reflects also my opinion on what could be a future role of these carnivals, to serve as a path for certification of scientific content parallel to the current scientific journals.
Given that there were so few submissions I added some links myself. Hopefully in the next editions we can get some more publicity and participation :). Talking about future editions, the second edition of Bio::Blogs will be hosted by Neil and we have now a full month to make something up in our blogs and submit the link to bioblogs{at}gmail{dot}com.
Conference Reports
I selected a blog post from Alf describing what was discussed in a recent conference dedicated to Data Webs. There is a lot of information about potential ways to deal with the increase of data submitted all over the web in many different formats. I remember seeing the advert for this conference and I was intrigued to see Philip Bourne, the editor-in-chief of PLoS Computational Biology, among the speakers. I see know that he is involved in publishing tools under development in PLoS.
Primers & Reviews
Stew from Flags and Lollipops sent in this link to a review on the use of bioinformatics to hunt for disease related genes. He highlights a series of tools and methods that can be used to prioritize candidate genes for experimental validation.
Neil, the next host of Bio::Blogs spent some time with the BioPerl package called Bio::Graphics. He dedicated a blog entry to explain how to create graphics for your results with this package. He gives examples on how to make graphic representations of sequences mapped with blast hits and phosphorylation sites.
Chris, a usual around Nodalpoint, nominated a post in Evolgen:
Evolgen has an interesting post about the relative importance (and interest in) cis and trans acting genetic variation in evo-devo. A lot of (computational) energy has thus far been expended in finding regulatory motifs close to genes (ie, within promoter regions), and conserved elements in non-coding sequences. Rather predictably, cis-acting variants have received the lion's share of attention, probably because they present a more tractable problem. The post deals with work from the evo-devo and comparative genomics fields, but these problems have also been attacked from within-species variation perspectives, particularly the genetics of gene expression. But that's next month's post...
Blog articles
I get to link to my last post. I present some very preliminary results on the influence of protein age on the likelihood of protein-protein interactions. Have fun pointing out all the likely flaws in reasoning and hopefully useful ways to build on it.
To wrap things up here is an announcement by Pierre of a possibly useful applet implementing a Genetic Programming Algorithm. If you ever wanted to play around with genetic programming you can have a go with his applet.
That is it for this month. It is a short Bio::Blogs but I hope you find some of these things useful. Don’t forget to submit the links for the next edition before the end of July. Neil will take up the editorial role for #2 in his blog. If you know of a nice symbol that we might use for Bio::Blogs sent it in as well.
The likelihood that two proteins interact might depend on the proteins' age
Abstract
It has been previously shown[1] that S. cerevisiae proteins preferentially interact with proteins of the same estimated likely time of origin. Using a similar approach but focusing on a less broad evolutionary time span I observed that the likelihood for protein interactions depends on the proteins’ age.
Methods and Results
Protein-protein interactions for S. cerevisiae were obtained from BIND, excluding any interactions derived from protein complexes. I considered only proteins that were represented in this interactome (i.e. with one or more interactions).
In order to create groups of S. cerevisiae proteins with different average age I used the reciprocal best blast hit method to determine the most likely ortholog in eleven other yeast species (see figure 1 for species names).
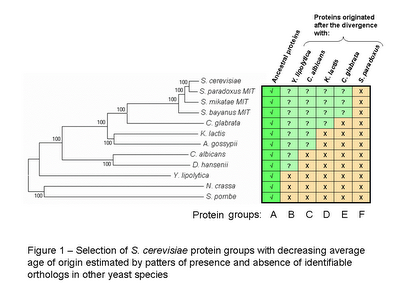
S. cerevisiae proteins with orthologs in all species were considered to be ancestral proteins and were grouped into group A. To obtain groups of proteins with decreasing average age of origin, S. cerevisiae proteins were selected according to the absence of identifiable orthologs in other species (see figure 1). It is important to note that these groups of decreasing average protein age are overlapping. Group F is contained in E , both are contained in D and so forth. I could have selected non overlapping groups of proteins with decreasing time of origin but the lower numbers obtained might in a latter stage make statistical analysis more difficult.
The phylogenetic tree in figure 1 (obtained with MEGA 3.1) is a neighbourhood joining tree obtained by concatenating 10 proteins from the ancestral group A. I did it mostly to avoid copyrighted images and too have a graphical representation of the species divergence.
To determine the effect of protein age on the likelihood of interaction with ancestral proteins I counted the number of interactions between group A and the other groups of proteins (see table 1).
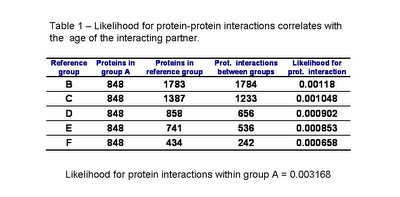
From the data it is possible to observe that protein-interactions within groups (within group A) is more likely than protein-interactions between groups. This is in agreement with the results from Qin et al.[1]. Also the likelihood for a protein to interact with an ancestral protein depends on the age of this protein. This simple analysis suggests that the younger the protein is the less likely it is to interact with an ancestral protein.
One possible use of this observation, if it holds to further scrutiny, would be to use the likely time of origin of the proteins as information to include in protein-protein prediction algorithms.
Caveats and possible continuations
The protein-protein interactions used here also contain the high-throughput studies and therefore the interactome used should be considered with caution. I might redo this analysis with a recent set of interactions compiled from the literature[2] but this will also introduce some bias into the interactome.
I should do some statistical analysis to determine if the differences observed are at all significant. If the differences are significant I should try to correlate the likelihood of interactions with a quantitative measure like average protein identity.
References
[1]Qin H, Lu HH, Wu WB, Li WH. Evolution of the yeast protein interaction network. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12820-4. Epub 2003 Oct 13
[2]Reguly T, Breitkreutz A, Boucher L, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006 Jun 8;5(4):11 [Epub ahead of print]
Abstract
It has been previously shown[1] that S. cerevisiae proteins preferentially interact with proteins of the same estimated likely time of origin. Using a similar approach but focusing on a less broad evolutionary time span I observed that the likelihood for protein interactions depends on the proteins’ age.
Methods and Results
Protein-protein interactions for S. cerevisiae were obtained from BIND, excluding any interactions derived from protein complexes. I considered only proteins that were represented in this interactome (i.e. with one or more interactions).
In order to create groups of S. cerevisiae proteins with different average age I used the reciprocal best blast hit method to determine the most likely ortholog in eleven other yeast species (see figure 1 for species names).

S. cerevisiae proteins with orthologs in all species were considered to be ancestral proteins and were grouped into group A. To obtain groups of proteins with decreasing average age of origin, S. cerevisiae proteins were selected according to the absence of identifiable orthologs in other species (see figure 1). It is important to note that these groups of decreasing average protein age are overlapping. Group F is contained in E , both are contained in D and so forth. I could have selected non overlapping groups of proteins with decreasing time of origin but the lower numbers obtained might in a latter stage make statistical analysis more difficult.
The phylogenetic tree in figure 1 (obtained with MEGA 3.1) is a neighbourhood joining tree obtained by concatenating 10 proteins from the ancestral group A. I did it mostly to avoid copyrighted images and too have a graphical representation of the species divergence.
To determine the effect of protein age on the likelihood of interaction with ancestral proteins I counted the number of interactions between group A and the other groups of proteins (see table 1).

From the data it is possible to observe that protein-interactions within groups (within group A) is more likely than protein-interactions between groups. This is in agreement with the results from Qin et al.[1]. Also the likelihood for a protein to interact with an ancestral protein depends on the age of this protein. This simple analysis suggests that the younger the protein is the less likely it is to interact with an ancestral protein.
One possible use of this observation, if it holds to further scrutiny, would be to use the likely time of origin of the proteins as information to include in protein-protein prediction algorithms.
Caveats and possible continuations
The protein-protein interactions used here also contain the high-throughput studies and therefore the interactome used should be considered with caution. I might redo this analysis with a recent set of interactions compiled from the literature[2] but this will also introduce some bias into the interactome.
I should do some statistical analysis to determine if the differences observed are at all significant. If the differences are significant I should try to correlate the likelihood of interactions with a quantitative measure like average protein identity.
References
[1]Qin H, Lu HH, Wu WB, Li WH. Evolution of the yeast protein interaction network. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12820-4. Epub 2003 Oct 13
[2]Reguly T, Breitkreutz A, Boucher L, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006 Jun 8;5(4):11 [Epub ahead of print]
Tags: protein interactions, evolution, networks
Sunday, June 25, 2006
Quick links
I stumbled upon a new computational biology blog called Nature's Numbers, looks interesting.
From Science Blogs universe here is a list compiled by Coturnix of upcoming blog carnivals for the next few days. I also remind anyone reading that the deadline for submissions for bio::blogs is coming very soon so send in your links :).
Still in Science Blogs here is an introduction to information theory. I am getting interesting in this as a tool for computational biology but I have a lot to learn on the subject. Here are two papers I fished out that use information theory in biology.
Also, if you want to donate some money, go check out the donors choose challenge of several Science Bloggers. Seed will match the donations up to $10,000 making each donation potentially more useful.
I stumbled upon a new computational biology blog called Nature's Numbers, looks interesting.
From Science Blogs universe here is a list compiled by Coturnix of upcoming blog carnivals for the next few days. I also remind anyone reading that the deadline for submissions for bio::blogs is coming very soon so send in your links :).
Still in Science Blogs here is an introduction to information theory. I am getting interesting in this as a tool for computational biology but I have a lot to learn on the subject. Here are two papers I fished out that use information theory in biology.
Also, if you want to donate some money, go check out the donors choose challenge of several Science Bloggers. Seed will match the donations up to $10,000 making each donation potentially more useful.
Wednesday, June 21, 2006
Journal policies on preprint servers
I mentioned in a previous post that it would be interesting to separate the registration, which allows claims of precedence for a scholarly finding (the submission of a manuscript) from the certification, which establishes the validity of a registered scholarly claim (the peer review process).
This can only happen if journals accept that a manuscript submitted to a preprint server is different from a peer-review article and therefore it should not be considered as prior publication. So what do the journals currently say about preprint servers ? I looked around the different policies, sent some emails and compiled a this list:
Nature: yes but ...
I enquired about this last part of their policy on the peer review forum and this was the response:
Nature Genetics/Nature Biotechnology: yes
PNAS: Yes!
BMC Bioinformatics/BMC Biology/BMC Evolutionary Biology/BMC Genomics/BMC Genetics/Genome Biology: Yes
Molecular Systems Biology: Do you feel lucky ?
Genome Research: No
Science: Do you feel lucky ?
Cell: No ?
PLoS - No clear policy information on the site about this but according to an email I got from PLoS they do consider for publication papers that have been submited in preprint servers. I hope they could make this clear in the policies they have available online.
Bioinformatics,Molecular Biology and Evolution - ??
I sent emails to both journals but I only had an answer from MBE directing me to this policy common to the journals of the Oxford University Press.
In summary most journals I checked will consider papers that have been previously submited to preprint servers, so I might consider in the future to submit my own work to preprint servers before looking for a journal. Very few journals clearly refuse manuscripts that might be available in electronic form but a good number either have no clear policy or reserve the right to reject papers that are available online.
I mentioned in a previous post that it would be interesting to separate the registration, which allows claims of precedence for a scholarly finding (the submission of a manuscript) from the certification, which establishes the validity of a registered scholarly claim (the peer review process).
This can only happen if journals accept that a manuscript submitted to a preprint server is different from a peer-review article and therefore it should not be considered as prior publication. So what do the journals currently say about preprint servers ? I looked around the different policies, sent some emails and compiled a this list:
Nature: yes but ...
Nature allows prior publication on recognised community preprint servers for review by other scientists in the field before formal submission to a journal. The details of the preprint server concerned and any accession numbers should be included in the cover letter accompanying submission of the manuscript to Nature. This policy does not extend to preprints available to the media or that are otherwise publicised before or during the submission and consideration process at Nature.
I enquired about this last part of their policy on the peer review forum and this was the response:
"We are aware that preprint servers such as ArXiv are available to the media, but as things stand we consider for publication, and publish, many papers that have been posted on it, and on other community preprint servers.As long as the authors have not actively sought out media coverage before submission and publication in Nature, we are happy to consider their work."
Nature Genetics/Nature Biotechnology: yes
(...)the presentation of results at scientific meetings (including the publication of abstracts) is acceptable, as is the deposition of unrefereed preprints in electronic archives.
PNAS: Yes!
"Preprints have a long and notable history in science, and it has been PNAS policy that they do not constitute prior publication. This is true whether an author hands copies of a manuscript to a few trusted colleagues or puts it on a publicly accessible web site for everyone to read, as is common now in parts of the physics community. The medium of distribution is not germane. A preprint is not considered a publication because it has not yet been formally reviewed and it is often not the final form of the paper. Indeed, a benefit of preprints is that feedback usually leads to an improved published paper or to no publication because of a revealed flaw. "
BMC Bioinformatics/BMC Biology/BMC Evolutionary Biology/BMC Genomics/BMC Genetics/Genome Biology: Yes
"Any manuscript or substantial parts of it submitted to the journal must not be under consideration by any other journal although it may have been deposited on a preprint server."
Molecular Systems Biology: Do you feel lucky ?
"Molecular Systems Biology reserves the right not to publish material that has already been pre-published (either in electronic or other media)."
Genome Research: No
"Submitted manuscripts must not be posted on any web site and are subject to press embargo."
Science: Do you feel lucky ?
"We will not consider any paper or component of a paper that has been published or is under consideration for publication elsewhere. Distribution on the Internet may be considered prior publication and may compromise the originality of the paper or submission. Please contact the editors with questions regarding allowable postings under this policy."
Cell: No ?
"Manuscripts are considered with the understanding that no part of the work has been published previously in print or electronic format and the paper is not under consideration by another publication or electronic medium."
PLoS - No clear policy information on the site about this but according to an email I got from PLoS they do consider for publication papers that have been submited in preprint servers. I hope they could make this clear in the policies they have available online.
Bioinformatics,Molecular Biology and Evolution - ??
"Authors wishing to deposit their paper in public or institutional repositories may deposit a link that provides free access to the paper, but must stipulate that public availability be delayed until 12 months after first online publication in the journal"
I sent emails to both journals but I only had an answer from MBE directing me to this policy common to the journals of the Oxford University Press.
In summary most journals I checked will consider papers that have been previously submited to preprint servers, so I might consider in the future to submit my own work to preprint servers before looking for a journal. Very few journals clearly refuse manuscripts that might be available in electronic form but a good number either have no clear policy or reserve the right to reject papers that are available online.
Tags: science, peer review, preprint servers
Monday, June 19, 2006
Mendel's Garden and Science Online Seminars
For those interested in evolution and genetics this is a good day. The first issue of Mendel's Garden is out with lots of interesting links. I particularly liked RPM's post on evolution of regulatory regions. I still think that evo-devo should focus a bit more on changes in protein interaction networks but more about that one of these days (hopefully :).
On a related note, Science started a series of online seminars with a primer on "Examining Natural Selection in Humans". This is a flash presentation with voice overs from the authors of a recent Science review on the same subject. I like this idea much more than the podcasts. I am not a big fan of podcasts because it is much faster to scan a text than it is to hear someone read it for you. At least with images there is more information and more appeal to spend some minutes listening to a presentation. The only thing I have against this Science Online Seminars initiative is that there is no RSS feed (I hope it is just a matter of time).
For those interested in evolution and genetics this is a good day. The first issue of Mendel's Garden is out with lots of interesting links. I particularly liked RPM's post on evolution of regulatory regions. I still think that evo-devo should focus a bit more on changes in protein interaction networks but more about that one of these days (hopefully :).
On a related note, Science started a series of online seminars with a primer on "Examining Natural Selection in Humans". This is a flash presentation with voice overs from the authors of a recent Science review on the same subject. I like this idea much more than the podcasts. I am not a big fan of podcasts because it is much faster to scan a text than it is to hear someone read it for you. At least with images there is more information and more appeal to spend some minutes listening to a presentation. The only thing I have against this Science Online Seminars initiative is that there is no RSS feed (I hope it is just a matter of time).
Tags: evolution, blog carnivals, science
Friday, June 16, 2006
Bio::Blogs announcement
Bio::Blogs is a blog carnival covering all bioinformatics and computational biology subjects. Bio::Blogs is schedule to be a monthly edition to come out on the first day of every month. The deadline for submission is until the end of month. Submissions for the next release of Bio::Blogs and offers to host the next editions can be sent to:
I will be hosting the first issue of Bio::Blogs here and there will be a homepage to keep track of all of the editions.
For discussions relating Bio::Blogs visit the Nodalpoint forum entry.
Bio::Blogs is a blog carnival covering all bioinformatics and computational biology subjects. Bio::Blogs is schedule to be a monthly edition to come out on the first day of every month. The deadline for submission is until the end of month. Submissions for the next release of Bio::Blogs and offers to host the next editions can be sent to:

I will be hosting the first issue of Bio::Blogs here and there will be a homepage to keep track of all of the editions.
For discussions relating Bio::Blogs visit the Nodalpoint forum entry.
Tags: blog carnivals, science, bioinformatics
Wednesday, June 14, 2006
SB2.0 webcast and other links
If you missed the Synthetic Biology 2.0 conference you can know watch the webcast here (via MainlyMartian).
Nature tech team over at Nascent continue with their productive stream of new products, including the release of Nature Network Boston and a new release of the Open Text Mining Interface. They even set up a webpage for us to keep up with all the activity here. They really look like a research group by know :) I wonder what will happen if they tried to publish some of this research... Open Text Mining Interface published by Nature in journal X.
If you missed the Synthetic Biology 2.0 conference you can know watch the webcast here (via MainlyMartian).
Nature tech team over at Nascent continue with their productive stream of new products, including the release of Nature Network Boston and a new release of the Open Text Mining Interface. They even set up a webpage for us to keep up with all the activity here. They really look like a research group by know :) I wonder what will happen if they tried to publish some of this research... Open Text Mining Interface published by Nature in journal X.
Monday, June 12, 2006
PLoS blogs
Liz Allen and Chris Surridge just kicked off the new PLoS blogs. According to Liz the blogs will be used to discuss their "vision for scientific communication, with all of its potentials and obstacles". I thank both of them for the nice links to this blog :) and for engaging in conversation.
Chris Surridge details in his first post how news of PLoS One has been spreading through the blogs. I think this only happened because the ideas behind ONE do strike a chord with bloggers and I really hope their efforts are met with success and more people engage in scientific discussion and collaboration.
Liz Allen and Chris Surridge just kicked off the new PLoS blogs. According to Liz the blogs will be used to discuss their "vision for scientific communication, with all of its potentials and obstacles". I thank both of them for the nice links to this blog :) and for engaging in conversation.
Chris Surridge details in his first post how news of PLoS One has been spreading through the blogs. I think this only happened because the ideas behind ONE do strike a chord with bloggers and I really hope their efforts are met with success and more people engage in scientific discussion and collaboration.
Science blog carnivals
What is a blog carnival ? In my opinion a blog carnival is just a meta-blog, a link aggregation supervised by an editor. They have been around for some time and there are already some rules to what usually is common to expect from a blog carnival. You can read this nice post on Science and Politics to get a better understanding of blog carnivals.
Here is a short summary I found on this FAQ:
There are of course science carnivals and I would say that their numbers are increasing with more people joining the science blogosphere. To my knowledge (please correct me :) the first scientific blog carnival was the Tangled Bank that I think started on the 21st of April 2004 and is still up and running.
These carnivals could also be seen as a path of certification (as discussed in the previous post). The rotating editor reviews submissions and bundles some of them together. This should guaranty that the carnival has the best of what has been posted on the subject in the recent past. The authors gain the attention of anyone interested in the carnival and the readers get supposably good quality posts on the subject. With time, and if there are more blog posts than carnivals we will likely see some carnivals gaining reputation.
Maybe one day having one of your discovery posts appear in one of the carnivals will be the equivalent of today having a paper published in a top journal.
With that said, why don't we start a computational biology/bioinformatics carnival ? :) There might not be enough people for it but we can make it monthly or something like this. Any suggestion for a name ?
What is a blog carnival ? In my opinion a blog carnival is just a meta-blog, a link aggregation supervised by an editor. They have been around for some time and there are already some rules to what usually is common to expect from a blog carnival. You can read this nice post on Science and Politics to get a better understanding of blog carnivals.
Here is a short summary I found on this FAQ:
Blog Carnivals typically collect together links pointing to blog articles on a particular topic. A Blog Carnival is like a magazine. It has a title, a topic, editors, contributors, and an audience. Editions of the carnival typically come out on a regular basis (e.g. every monday, or on the first of the month). Each edition is a special blog article that consists of links to all the contributions that have been submitted, often with the editors opinions or remarks.
There are of course science carnivals and I would say that their numbers are increasing with more people joining the science blogosphere. To my knowledge (please correct me :) the first scientific blog carnival was the Tangled Bank that I think started on the 21st of April 2004 and is still up and running.
These carnivals could also be seen as a path of certification (as discussed in the previous post). The rotating editor reviews submissions and bundles some of them together. This should guaranty that the carnival has the best of what has been posted on the subject in the recent past. The authors gain the attention of anyone interested in the carnival and the readers get supposably good quality posts on the subject. With time, and if there are more blog posts than carnivals we will likely see some carnivals gaining reputation.
Maybe one day having one of your discovery posts appear in one of the carnivals will be the equivalent of today having a paper published in a top journal.
With that said, why don't we start a computational biology/bioinformatics carnival ? :) There might not be enough people for it but we can make it monthly or something like this. Any suggestion for a name ?
Thursday, June 08, 2006
The peer review trial
The next day after finding about PLoS One I saw the announcement for the Nature peer review trial. For the next couple of months any author submitting to Nature can opt to go trough a parallel process of open peer review. Nature is also promoting the discussion on the issue online in a forum where anyone can comment. You can also track the discussion going on the web through Connotea under the tag of "peer review trial", or under the "peer review" tag in Postgenomic.
I really enjoyed reading this opinion on "Rethinking Scholarly Communication", summarized in one of the Nature articles. Briefly, the authors first describe (from Roosendaal and Geurts) the required functions any system of scholarly communication:
* Registration, which allows claims of precedence for a scholarly finding.
* Certification, which establishes the validity of a registered scholarly claim.
* Awareness, which allows actors in the scholarly system to remain aware of new claims and findings.
* Archiving, which preserves the scholarly record over time.
* Rewarding, which rewards actors for their performance in the communication system based on metrics derived from that system.
The authors then try to show that it is possible to build a science communication system where all these functions are not centered in the journal, but are separated in different entities.
This would speed up science communication. There is a significant delay between submitting a communication and having it accessible to others because all the functions are centered in the journals and only after the certification (peer reviewing) is the work made available.
Separating the registration from the certification also has the potential benefit of exploring parallel certifications. The manuscripts deposited in the pre-print servers can be evaluated by the traditional peer-review process in journals but on top of this there is also the possibility of exploring other ways of certifying the work presented. The authors give the example of Citabase but also blog aggregation sites like Postgenomic could provide independent measures of the interest of a communication.
More generally and maybe going a bit of-topic, this reminded me of the correlation between modularity and complexity in biology. By dividing a process into separate and independent modules you allow for exploration of novelty without compromising the system. The process is still free to go from start to end in the traditional way but new subsystems can be created to compete with some of modules.
For me this discussion, is relevant for the whole scientific process , not just communication. New web technologies lower the costs of establishing collaborations and should therefore ease the recruitment of resources required to tackle a problem. Because people are better at different task it does make some sense to increase the modularity in the scientific process.
The next day after finding about PLoS One I saw the announcement for the Nature peer review trial. For the next couple of months any author submitting to Nature can opt to go trough a parallel process of open peer review. Nature is also promoting the discussion on the issue online in a forum where anyone can comment. You can also track the discussion going on the web through Connotea under the tag of "peer review trial", or under the "peer review" tag in Postgenomic.
I really enjoyed reading this opinion on "Rethinking Scholarly Communication", summarized in one of the Nature articles. Briefly, the authors first describe (from Roosendaal and Geurts) the required functions any system of scholarly communication:
* Registration, which allows claims of precedence for a scholarly finding.
* Certification, which establishes the validity of a registered scholarly claim.
* Awareness, which allows actors in the scholarly system to remain aware of new claims and findings.
* Archiving, which preserves the scholarly record over time.
* Rewarding, which rewards actors for their performance in the communication system based on metrics derived from that system.
The authors then try to show that it is possible to build a science communication system where all these functions are not centered in the journal, but are separated in different entities.
This would speed up science communication. There is a significant delay between submitting a communication and having it accessible to others because all the functions are centered in the journals and only after the certification (peer reviewing) is the work made available.
Separating the registration from the certification also has the potential benefit of exploring parallel certifications. The manuscripts deposited in the pre-print servers can be evaluated by the traditional peer-review process in journals but on top of this there is also the possibility of exploring other ways of certifying the work presented. The authors give the example of Citabase but also blog aggregation sites like Postgenomic could provide independent measures of the interest of a communication.
More generally and maybe going a bit of-topic, this reminded me of the correlation between modularity and complexity in biology. By dividing a process into separate and independent modules you allow for exploration of novelty without compromising the system. The process is still free to go from start to end in the traditional way but new subsystems can be created to compete with some of modules.
For me this discussion, is relevant for the whole scientific process , not just communication. New web technologies lower the costs of establishing collaborations and should therefore ease the recruitment of resources required to tackle a problem. Because people are better at different task it does make some sense to increase the modularity in the scientific process.
Tags: peer review, peer review trial, science2.0
Monday, June 05, 2006
PLoS One
There is an article in Wired about open access in scientific publishing. It focuses on the efforts of the Public library of Science (PLoS) to make content freely available by transferring the costs of publication to the authors. What actually caught my attention was this little paragraph:
The success of the top two PLoS journals has led to the birth of four more modest ones aimed at specific fields: clinical trials, computational biology, genetics, and pathogens. And this summer, Varmus and his colleagues will launch PLoS One, a paperless journal that will publish online any paper that evaluators deem “scientifically legitimate.” Each article will generate a thread for comment and review. Great papers will be recognized by the discussion they generate, and bad ones will fade away.
The emphasis is mine. I went snooping around for the upcoming PLoS One and I found a page to subscribe to a mailing list. It has curious banner with a subtitle of open access 2.0.

I found some links in the source code that got me to the prototype webpage. It sounds exactly like what a lot of people have been pushing for: rapid scientific communication, community peer reviewing, continuous revision of the paper (they call it interactive papers) and open access. This will be hard to implement but if successful it will do much to bring more transparency to the scientific process and increase the cooperation between scientist.
There is also something about the name PLoS ONE. They are really betting a lot on this launch if they are calling it ONE. It implicitly states that ONE will be the flagship of PLoS, where any paper (not just Biology) can be published.
There is an article in Wired about open access in scientific publishing. It focuses on the efforts of the Public library of Science (PLoS) to make content freely available by transferring the costs of publication to the authors. What actually caught my attention was this little paragraph:
The success of the top two PLoS journals has led to the birth of four more modest ones aimed at specific fields: clinical trials, computational biology, genetics, and pathogens. And this summer, Varmus and his colleagues will launch PLoS One, a paperless journal that will publish online any paper that evaluators deem “scientifically legitimate.” Each article will generate a thread for comment and review. Great papers will be recognized by the discussion they generate, and bad ones will fade away.
The emphasis is mine. I went snooping around for the upcoming PLoS One and I found a page to subscribe to a mailing list. It has curious banner with a subtitle of open access 2.0.

I found some links in the source code that got me to the prototype webpage. It sounds exactly like what a lot of people have been pushing for: rapid scientific communication, community peer reviewing, continuous revision of the paper (they call it interactive papers) and open access. This will be hard to implement but if successful it will do much to bring more transparency to the scientific process and increase the cooperation between scientist.
There is also something about the name PLoS ONE. They are really betting a lot on this launch if they are calling it ONE. It implicitly states that ONE will be the flagship of PLoS, where any paper (not just Biology) can be published.
Tags: open access, PLoS, PLoS One, science, science 2.0
Wednesday, May 31, 2006
Bringing democracy to the net
Democracy is most often thought of as in opposition to totalitarianism. In this case I mean democracy in opposition to anarchy. Some people are raising their voices against the trend of collective intelligence/wisdom of the crows that have been the hype of the net for the past few years. Wikipedia is the crown jewel of this trend of empowering people for the common good and probably as a result of the project's visibility it has been the one to take the heat from the backlash.
Is Wikipedia dead like Nicholas Carr suggests in his blog ? His provocative title was a flame bait but he does call attention to some interesting things happening at Wikipedia. The wikipedia is not dead , it is just changing. It has to change to cope with the increase in visibility, vandalism and to deal with situations where no real consensus is possible.
The system is evolving by restricting anonymous posting and allowing editors to apply temporary editing restrictions to some pages. It is evolving to become more bureaucratic in nature with disputes and mechanisms to deal with the discord. What Nicholas Carr said is dead is the ideal that anyone can edit anything in wikipedia and I would say this is actually good news.
Following his post on the death of Wikipedia, Carr points to an assay by Jaron Lanier entitled Digital Maoism. It is a bit long but I highly recommend it.
Some quotes from the text:
"Every authentic example of collective intelligence that I am aware of also shows how that collective was guided or inspired by well-meaning individuals. These people focused the collective and in some cases also corrected for some of the common hive mind failure modes. The balancing of influence between people and collectives is the heart of the design of democracies, scientific communities, and many other long-standing projects. "
Sites like Wikipedia are important online experiments. They are trying to develop the tools that allow useful work to come out from millions of very small contributions. I think this will have to go trough some representative democracy systems. We still have to work on ways to establish the governing body in these internet systems. Essentially to whom we decide to deposit trust for that particular task or realm of knowledge. For this we will need better ways to define identity online and to establish trust relationships.
Further reading:
Wiki-truth
Democracy is most often thought of as in opposition to totalitarianism. In this case I mean democracy in opposition to anarchy. Some people are raising their voices against the trend of collective intelligence/wisdom of the crows that have been the hype of the net for the past few years. Wikipedia is the crown jewel of this trend of empowering people for the common good and probably as a result of the project's visibility it has been the one to take the heat from the backlash.
Is Wikipedia dead like Nicholas Carr suggests in his blog ? His provocative title was a flame bait but he does call attention to some interesting things happening at Wikipedia. The wikipedia is not dead , it is just changing. It has to change to cope with the increase in visibility, vandalism and to deal with situations where no real consensus is possible.
The system is evolving by restricting anonymous posting and allowing editors to apply temporary editing restrictions to some pages. It is evolving to become more bureaucratic in nature with disputes and mechanisms to deal with the discord. What Nicholas Carr said is dead is the ideal that anyone can edit anything in wikipedia and I would say this is actually good news.
Following his post on the death of Wikipedia, Carr points to an assay by Jaron Lanier entitled Digital Maoism. It is a bit long but I highly recommend it.
Some quotes from the text:
"Every authentic example of collective intelligence that I am aware of also shows how that collective was guided or inspired by well-meaning individuals. These people focused the collective and in some cases also corrected for some of the common hive mind failure modes. The balancing of influence between people and collectives is the heart of the design of democracies, scientific communities, and many other long-standing projects. "
Sites like Wikipedia are important online experiments. They are trying to develop the tools that allow useful work to come out from millions of very small contributions. I think this will have to go trough some representative democracy systems. We still have to work on ways to establish the governing body in these internet systems. Essentially to whom we decide to deposit trust for that particular task or realm of knowledge. For this we will need better ways to define identity online and to establish trust relationships.
Further reading:
Wiki-truth
Tags: wikipedia, web2.0, collective intelligence
Friday, May 26, 2006
The Human Puppet (2)
In November I rambled about a possible sci-fi scenario. It was about a human person giving away their will to be directed by the masses in the internet. A vessel for the "collective intelligence". A voluntary and extreme reality show.
Well, there goes the sci-fi, you can participate in it in about 19 days. Via TechCrunch I found this site:
Kieran Vogel will make Internet television history when he becomes the first person to give total control of his life to the Internet.
(...)
Through an interactive media platform Kieran will live by the decisions the internet decides such as:
# What time he wakes up
# What he wears
# What he eats
# Who he dates
# What he watches
I get a visceral negative response to this. Although this is just a reality show and it is all going to happen inside a house I think it will be important to keep this in mind. In the future technology will make web even more pervasive then today and there are scenarios along the lines of this human puppet idea that could have negative consequences.
I guess what I am thinking is that the same technologies that helps us to collaborate can also be use to control (sounds a bit obvious). In the end the only difference is on how much do the people involved want to (or can) exercise their will power.
In November I rambled about a possible sci-fi scenario. It was about a human person giving away their will to be directed by the masses in the internet. A vessel for the "collective intelligence". A voluntary and extreme reality show.
Well, there goes the sci-fi, you can participate in it in about 19 days. Via TechCrunch I found this site:
Kieran Vogel will make Internet television history when he becomes the first person to give total control of his life to the Internet.
(...)
Through an interactive media platform Kieran will live by the decisions the internet decides such as:
# What time he wakes up
# What he wears
# What he eats
# Who he dates
# What he watches
I get a visceral negative response to this. Although this is just a reality show and it is all going to happen inside a house I think it will be important to keep this in mind. In the future technology will make web even more pervasive then today and there are scenarios along the lines of this human puppet idea that could have negative consequences.
I guess what I am thinking is that the same technologies that helps us to collaborate can also be use to control (sounds a bit obvious). In the end the only difference is on how much do the people involved want to (or can) exercise their will power.
Thursday, May 25, 2006
Using viral memes to request computer time
Every time we direct the browser somewhere, dedicating your attention, some computer processing time is used to display the page. This includes a lot of client side processing like all the javascript in all that nice looking AJAX stuff. What if we could harvest some of this computer processing power to solve very small tasks, something like grid computing.
How would this work ? There could be a video server that would allow me to put a video on my blog (like google video) or a simple game or whatever thing that people would enjoy and spend a little time doing. During this time there would be a package downloaded from the same server, some processing done on the client side and a result sent back. If people enjoy the video/game/whatever and it goes viral then it spreads all over the blogs and any person dedicating their attention to it is contributing computer power to solve some task. Maybe this could work as an alternative to advertising ? Good content would be traded for computer power. To compare, Sun is selling computer power in the US for 1 dollar an hour. Of course this type of very small scale grid processing would be worth much less.
Every time we direct the browser somewhere, dedicating your attention, some computer processing time is used to display the page. This includes a lot of client side processing like all the javascript in all that nice looking AJAX stuff. What if we could harvest some of this computer processing power to solve very small tasks, something like grid computing.
How would this work ? There could be a video server that would allow me to put a video on my blog (like google video) or a simple game or whatever thing that people would enjoy and spend a little time doing. During this time there would be a package downloaded from the same server, some processing done on the client side and a result sent back. If people enjoy the video/game/whatever and it goes viral then it spreads all over the blogs and any person dedicating their attention to it is contributing computer power to solve some task. Maybe this could work as an alternative to advertising ? Good content would be traded for computer power. To compare, Sun is selling computer power in the US for 1 dollar an hour. Of course this type of very small scale grid processing would be worth much less.
Wednesday, May 24, 2006
Conference blogging and SB2.0
In case you missed the Synthetic Biology 2.0 meeting and want a quick summary of what happened there you can take a look at some blogs. There were at least 4 bloggers at the conference. Oliver Morton (chief news and features editor of Nature) has a series of posts in his old blog. Rob Carlson described how he and Drew Endy were calling the field intentional biology. Alex Mallet from Drew Endy's lab has a quick summary of the meeting and finally Mackenzie has in his cis-action by far the best coverage with lots more to read.
I hope they put up on the site the recorded talks since I missed a lot of interesting things during the live webcast.
In the third day of the meeting (that was not available in the live webcast) there was a discussion about possible self-regulation in the field (as in the 1975 Asilomar meeting). According to an article in NewScientist the attending researchers decided against self-regulation measures.
In case you missed the Synthetic Biology 2.0 meeting and want a quick summary of what happened there you can take a look at some blogs. There were at least 4 bloggers at the conference. Oliver Morton (chief news and features editor of Nature) has a series of posts in his old blog. Rob Carlson described how he and Drew Endy were calling the field intentional biology. Alex Mallet from Drew Endy's lab has a quick summary of the meeting and finally Mackenzie has in his cis-action by far the best coverage with lots more to read.
I hope they put up on the site the recorded talks since I missed a lot of interesting things during the live webcast.
In the third day of the meeting (that was not available in the live webcast) there was a discussion about possible self-regulation in the field (as in the 1975 Asilomar meeting). According to an article in NewScientist the attending researchers decided against self-regulation measures.
Saturday, May 20, 2006
Synthetic Biology & best practices
There is a Synthetic Biology conference going on in Berkeley (webcast here) and they are going to talk about the subject of best practices in one of the days. There is a document online with an outline of some of the subjects up for discussion. In reaction to this, a group of organization published an open letter for the people attending the meeting.
From the text:
We are writing to express our deep concerns about the rapidly developing field of Synthetic Biology that is attempting to create novel life forms and artificial living systems. We believe that this potentially powerful technology is being developed without proper societal debate concerning socio-economic, security, health, environmental and human rights implications. We are alarmed that synthetic biologists meeting this weekend intend to vote on a scheme of voluntary self-regulation without consulting or involving broader social groups. We urge you to withdraw these self-governance proposals and participate in a process of open and inclusive oversight of this technology.
Forms of self-regulation are not incompatible with open discussion with the broader society nor with state regulation. Do we even need regulation at this point ?
There is a Synthetic Biology conference going on in Berkeley (webcast here) and they are going to talk about the subject of best practices in one of the days. There is a document online with an outline of some of the subjects up for discussion. In reaction to this, a group of organization published an open letter for the people attending the meeting.
From the text:
We are writing to express our deep concerns about the rapidly developing field of Synthetic Biology that is attempting to create novel life forms and artificial living systems. We believe that this potentially powerful technology is being developed without proper societal debate concerning socio-economic, security, health, environmental and human rights implications. We are alarmed that synthetic biologists meeting this weekend intend to vote on a scheme of voluntary self-regulation without consulting or involving broader social groups. We urge you to withdraw these self-governance proposals and participate in a process of open and inclusive oversight of this technology.
Forms of self-regulation are not incompatible with open discussion with the broader society nor with state regulation. Do we even need regulation at this point ?
Tags: conference, Synthetic Biology
The internet and the study of human intelligence
I started reading a book on machine learning methods last night and my mind floated away to thinking about the internet and artificial intelligence (yes the book is a bit boring :).
Anyway, one thing that I thought about was how the internet might become (or is already) a very good place to study (human) intelligence. Some people are very transparent on the net and if anything the trend is for people to start sharing their lives or at least their view of the world earlier. So it is possible to get an idea of what someone is exposed to, what people read, films they see, some of their life experiences, etc. In some sense you can access someone's input in life.
On the other hand you can also read this person's opinions when presented with some content. Person X with known past experiences Y was exposed to Z and reacted in this way. With this information we could probably learn a lot about human thought processes.
I started reading a book on machine learning methods last night and my mind floated away to thinking about the internet and artificial intelligence (yes the book is a bit boring :).
Anyway, one thing that I thought about was how the internet might become (or is already) a very good place to study (human) intelligence. Some people are very transparent on the net and if anything the trend is for people to start sharing their lives or at least their view of the world earlier. So it is possible to get an idea of what someone is exposed to, what people read, films they see, some of their life experiences, etc. In some sense you can access someone's input in life.
On the other hand you can also read this person's opinions when presented with some content. Person X with known past experiences Y was exposed to Z and reacted in this way. With this information we could probably learn a lot about human thought processes.
A little bit of this a little bit of that ...
What do you get when you mix humans/sex/religion/evolution? A big media hype.
Also, given that a big portion of the scientist currently blogging are working on evolution you also get a lot of buzzing in the science blogosphere. No wonder then why this paper reached the top spot in postgenomic.
This one is a very good example of the usefulness of blogs and why we should really promote more science communication online. The paper was released in advanced online publication and some days after you can already read a lot of opinions about it. It is not just the blog entries but also all the comments on these blog posts. As a result of this we not only get the results and discussion from the paper but the opinion of whoever decided to participate in the discussion.
What do you get when you mix humans/sex/religion/evolution? A big media hype.
Also, given that a big portion of the scientist currently blogging are working on evolution you also get a lot of buzzing in the science blogosphere. No wonder then why this paper reached the top spot in postgenomic.
This one is a very good example of the usefulness of blogs and why we should really promote more science communication online. The paper was released in advanced online publication and some days after you can already read a lot of opinions about it. It is not just the blog entries but also all the comments on these blog posts. As a result of this we not only get the results and discussion from the paper but the opinion of whoever decided to participate in the discussion.
Tags: science, evolution, science communication
Wednesday, May 17, 2006
Postgenomic greasemonkey script (2)
I have posted the Postgenomic script I mentioned in the previous post in the Nodalpoint wiki page. There are some instructions there on how to get it running. If you have some problems or suggestions leave some comments here or in the forum in Nodalpoint. Right now it is only set to work with the Nature journals but it should work with more.
I have posted the Postgenomic script I mentioned in the previous post in the Nodalpoint wiki page. There are some instructions there on how to get it running. If you have some problems or suggestions leave some comments here or in the forum in Nodalpoint. Right now it is only set to work with the Nature journals but it should work with more.
Tags: postgenomic
Saturday, May 13, 2006
Postgenomics script for Firefox
I am playing around with greasemonkey to try to add links to Postgenomic to journal websites. The basic idea is to search the webpage you are seeing (like a Nature website for example) for papers that have been talked about in blogs and are tracked by Postgenomic. When one is found a little picture is added with a link to the Postgenomic page talking about the paper.
The result is something like this (in the case of the table of contents):

Or like this when viewing the paper itself:

In another journal:

I am more comfortable with Perl, but anyway I think it works as a proof or principle. If Stew agrees I'll probably post the script in Nodalpoint for people to improve or just try it out.
I am playing around with greasemonkey to try to add links to Postgenomic to journal websites. The basic idea is to search the webpage you are seeing (like a Nature website for example) for papers that have been talked about in blogs and are tracked by Postgenomic. When one is found a little picture is added with a link to the Postgenomic page talking about the paper.
The result is something like this (in the case of the table of contents):

Or like this when viewing the paper itself:

In another journal:

I am more comfortable with Perl, but anyway I think it works as a proof or principle. If Stew agrees I'll probably post the script in Nodalpoint for people to improve or just try it out.
Tags: postgenomic, blogs
Thursday, May 11, 2006
Google Trends and Co-Op
There some new Google services up and running and buzzing around the blogs today. I only briefly took a look around them.
Google Trends is like Google Finance for anything search trend than you want to analyze. Very useful for someone wanting to waste time instead of doing some productive work ;). You can compare the search and news volume for different terms like:
It gets the data from all the google searches so it really does not reflect the trends within the scientific community.
The other new tool out yesterday is Google Co-Op, the start of social search for Google. It looks as obscure as Google Base so I can again try to make some weird connection to how researcher might use it :). It looks like Google Co-Op is a way for users to further personalize their search. User can subscribe to providers that offer their knowledge/guidance to shape some of the results you see in your search. If you search for example for alzheimer's you should see on the top of the results some refinement that you can do. For example you can look only at treatment related results. This was possible because a list of contributors have labeled a lot of content according to some rules.
Anyone can create a directory and start labeling content following an XML schema that describes the "context". So anyone or (more likely) any group of people can add metadata to content and have it available in google. The obvious application for science would be to have metadata on scientific publications available. Maybe getting Connotea and CiteULike data into a google directory for example would be useful. These sites can still go on developing the niche specific tools but we could benefit from having a lot of the tagging metadata available in google.
There some new Google services up and running and buzzing around the blogs today. I only briefly took a look around them.
Google Trends is like Google Finance for anything search trend than you want to analyze. Very useful for someone wanting to waste time instead of doing some productive work ;). You can compare the search and news volume for different terms like:

It gets the data from all the google searches so it really does not reflect the trends within the scientific community.
The other new tool out yesterday is Google Co-Op, the start of social search for Google. It looks as obscure as Google Base so I can again try to make some weird connection to how researcher might use it :). It looks like Google Co-Op is a way for users to further personalize their search. User can subscribe to providers that offer their knowledge/guidance to shape some of the results you see in your search. If you search for example for alzheimer's you should see on the top of the results some refinement that you can do. For example you can look only at treatment related results. This was possible because a list of contributors have labeled a lot of content according to some rules.
Anyone can create a directory and start labeling content following an XML schema that describes the "context". So anyone or (more likely) any group of people can add metadata to content and have it available in google. The obvious application for science would be to have metadata on scientific publications available. Maybe getting Connotea and CiteULike data into a google directory for example would be useful. These sites can still go on developing the niche specific tools but we could benefit from having a lot of the tagging metadata available in google.
Wednesday, May 10, 2006
Nature Protocols
Nature continues clearly the most innovative of the publishing houses in my view. A new web site is up in beta phase called Nature Protocols:
Nature Protocols is a new online web resource for laboratory protocols. The site, currently in beta phase, will contain high quality, peer-reviewed protocols commissioned by the Nature Protocols Editorial team and will also publish content posted onto the site by the community
They accept different types of content:
* Peer-reviewed protocols
* Protocols related to primary research papers in Nature journals
* Company Protocols and Application notes
* Non peer-reviewed (Community) protocols
There are already several protocol websites already out there so what is the point ? For Nature I guess it is obvious. Just like most portal websites they are creating a very good place to put ads. I am sure that all these protocols will have links to products on their Nature products and a lot of ads. The second advantage for Nature is the stickiness of the service. More people will come back to the website to look for protocols and stumble on to Nature content, increasing visibility for the journals and their impact.
A little detail is that, as they say above, the protocols from the papers published in the Nature journals will be made available on the website. On one hand this sounds great because the methods sections in the papers are usually so small (due to restrictions for publication) that they are most of the times incredibly hard to decipher (and usually put into supplementary materials). On the other hand, this will increase even further the tendency to hide away from the paper the really important pars of the research, the results and how these where obtained (methods) and to show only the subjective interpretations of the authors.
This reminds me of a recent editorial by Gregory A Petsko in Genome Biology (sub only). Here is how is states the problem :) - "The tendency to marginalize the methods is threatening to turn papers in journals like Nature and Science into glorified press releases."
For scientists this will be a very useful resource. Nature has a lot of appeal and will be able to quickly create a lot of really good content by inviting experienced scientists to write up their protocols full with tips and tricks accumulated over years of experience. This is the easy part for science portals, the content comes free. If somebody went to Yahoo and told them that scientist actually pay scientific journals to please please show our created content they would probably laugh :). Yahoo/MSN and other web portals have to pay people to create the content that they have on their sites.
Nature continues clearly the most innovative of the publishing houses in my view. A new web site is up in beta phase called Nature Protocols:
Nature Protocols is a new online web resource for laboratory protocols. The site, currently in beta phase, will contain high quality, peer-reviewed protocols commissioned by the Nature Protocols Editorial team and will also publish content posted onto the site by the community
They accept different types of content:
* Peer-reviewed protocols
* Protocols related to primary research papers in Nature journals
* Company Protocols and Application notes
* Non peer-reviewed (Community) protocols
There are already several protocol websites already out there so what is the point ? For Nature I guess it is obvious. Just like most portal websites they are creating a very good place to put ads. I am sure that all these protocols will have links to products on their Nature products and a lot of ads. The second advantage for Nature is the stickiness of the service. More people will come back to the website to look for protocols and stumble on to Nature content, increasing visibility for the journals and their impact.
A little detail is that, as they say above, the protocols from the papers published in the Nature journals will be made available on the website. On one hand this sounds great because the methods sections in the papers are usually so small (due to restrictions for publication) that they are most of the times incredibly hard to decipher (and usually put into supplementary materials). On the other hand, this will increase even further the tendency to hide away from the paper the really important pars of the research, the results and how these where obtained (methods) and to show only the subjective interpretations of the authors.
This reminds me of a recent editorial by Gregory A Petsko in Genome Biology (sub only). Here is how is states the problem :) - "The tendency to marginalize the methods is threatening to turn papers in journals like Nature and Science into glorified press releases."
For scientists this will be a very useful resource. Nature has a lot of appeal and will be able to quickly create a lot of really good content by inviting experienced scientists to write up their protocols full with tips and tricks accumulated over years of experience. This is the easy part for science portals, the content comes free. If somebody went to Yahoo and told them that scientist actually pay scientific journals to please please show our created content they would probably laugh :). Yahoo/MSN and other web portals have to pay people to create the content that they have on their sites.
Tags: Nature, science portals
web2.0@EMBL
The EMBL Centre for Computational Biology has announced a series of talks related to novel concepts and easy-to-use web tools for biologists. So far there are three schedule talks:

Session 1 - Using new web concepts for more efficient research - an introduction for the less-techy crowd
Time/place: Tue, May 16th, 2006; 14:30; Small Operon
This one I think will introduce the concepts around what is called web2.0 and the potential impact these might have for researchers. I am really curious to see how big will the "less-techy crowd" really be :).
The following sessions are a bit more specific dealing with particular problems we might have in our activities and how can some of the recent web technologies help us deal with them.
Session 2 - Information overflow? Stay tuned with a click (May 23rd, 2006; 14:30;)
Session 3 - Tags: simply organize and share links and references with keywords (May 30th, 2006; 14:30)
Session 4 - Stop emailing huge files: How to jointly edit manuscripts and share data (June 6th, 2006; 14:30;)
All in the Small Operon, here in the EMBL Heidelberg
I commend the efforts of the EMBL CCB and I hope that a lot of people turn up. Let's see if the open collaborative ideas come up on the discussions. If you are in the neighborhood and are interested, come on by and help with the discussion (map).
The EMBL Centre for Computational Biology has announced a series of talks related to novel concepts and easy-to-use web tools for biologists. So far there are three schedule talks:

Session 1 - Using new web concepts for more efficient research - an introduction for the less-techy crowd
Time/place: Tue, May 16th, 2006; 14:30; Small Operon
This one I think will introduce the concepts around what is called web2.0 and the potential impact these might have for researchers. I am really curious to see how big will the "less-techy crowd" really be :).
The following sessions are a bit more specific dealing with particular problems we might have in our activities and how can some of the recent web technologies help us deal with them.
Session 2 - Information overflow? Stay tuned with a click (May 23rd, 2006; 14:30;)
Session 3 - Tags: simply organize and share links and references with keywords (May 30th, 2006; 14:30)
Session 4 - Stop emailing huge files: How to jointly edit manuscripts and share data (June 6th, 2006; 14:30;)
All in the Small Operon, here in the EMBL Heidelberg
I commend the efforts of the EMBL CCB and I hope that a lot of people turn up. Let's see if the open collaborative ideas come up on the discussions. If you are in the neighborhood and are interested, come on by and help with the discussion (map).
Subscribe to:
Comments (Atom)