SyntheticBiology@Nature.comThis week Nature has a
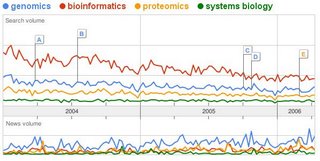
special issue on <buzz>Synthetic Biology</buzz>. I have currently a kind of love/hate relationship with trends in biology. It is easy to track the trends (in the recent past: genomics, proteomics, bioinformatics, systems biology, nanotechnology, synthetic biology) and it is somehow fascinating to follow them and watch them propagate. It holds for me a similar fascination has seeing a meme propagate in the web. Someone will still write a thesis on how a kid was able to put up a webpage
like this one and make truck load of money selling pixels just because he ignited the curiosity of people on a global scale.
There is always a reason behind each rising trend in biology, but they are clearly too short lived to deliver on their expectations, so what is the point ? Why do these waves of buzz exist in research ? The mentality of engineering in biology is not new so why the recent interest in synthetic biology ?
I am too young to know if this has always been like this but I am inclined to think that this is just to product of increasing competition for resources (grant applications). Every once in a while scientist have to re-invent the pressing reasons why society has to invest in them. The big projects that will galvanize the masses, the next genome project.
I personally like the engineering approach to biology. Much of the work that is done in the lab where I am doing my phd is engineering oriented. Synthetic biology (or whatever it was called in the past and will be called in the future) could deliver things like cheap energy (biological solar panels),cheaper chemicals (optimized systems of production), cheap food (GMOs or some even weirder tissue cultures), clean water, improved immune-systems, etc. A quick look at the two reviews that are in this week's issue of Nature will tell you that we are still far from all of this.
The review by David Sprinzak and Michael Elowitz tries to cover broadly what as been achieved in engineering biological systems in the last couple of years (references range from 2000 to 2005). Apart from the reference to
a paper on the engineering a mevalonate pathway in Escherichia coli, most of the work so far done in the field is preliminary. People have been trying to assemble simple systems and end up learning new things along the way.
The second review is authored by Drew Endy and is basically synthetic biology
evangelism :). Drew Endy has been one of the voices
shouting louder in support of this field and in looking for
standardization and
open exchange of information and materials (some
notes from the biomass). The only new thing he says in this review that I have not heard before from him is a short paragraph on evolution. We are used to engineering things that do not replicate (cars, computers, tv sets, etc) and the field will have to start thinking of the consequences of evolution of the systems it tinkers with. Are the systems sustainable ? Will they change within their useful life time ?
There is one accompanying
research paper reporting on a chimeric light sensing protein that is de-phosphorylated in the presence of red light. The bacteria produce lacZ in the dark and the production is decreased with increasing amounts of red light. You can make
funny pictures with these bacteria but has for the real scientific value of this discovery I can link to
two comments in Slashdot. Maybe that is exaggerated. Making chimeric protein receptors that work can be tricky and it is very nice that something started by
college students can end up in a Nature paper.
Last but not least there is a
comic ! The fantastic: "Adventures in Synthetic Biology". Ok, here is where I draw the line :) Who is this for ? Since when do teens read Nature ? How would they have access to this ? I like comics, I do ... but this is clearly not properly targeted.